Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
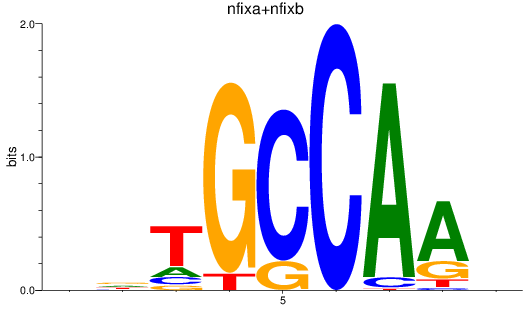
Results for nfixa+nfixb
Z-value: 0.85
Transcription factors associated with nfixa+nfixb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfixa
|
ENSDARG00000043226 | nuclear factor I/Xa |
|
nfixb
|
ENSDARG00000061836 | nuclear factor I/Xb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfixa | dr11_v1_chr1_+_51896147_51896147 | -0.32 | 1.9e-01 | Click! |
| nfixb | dr11_v1_chr3_+_14768364_14768364 | -0.29 | 2.4e-01 | Click! |
Activity profile of nfixa+nfixb motif
Sorted Z-values of nfixa+nfixb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_33645967 | 1.93 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr18_+_618005 | 1.88 |
ENSDART00000189667
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr11_-_11575070 | 1.58 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr8_-_22965214 | 1.28 |
ENSDART00000148178
|
emilin3a
|
elastin microfibril interfacer 3a |
| chr3_+_49021079 | 1.27 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr12_-_16595177 | 1.16 |
ENSDART00000133962
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr19_+_25465025 | 1.03 |
ENSDART00000018553
|
rpa3
|
replication protein A3 |
| chr4_+_25680480 | 1.03 |
ENSDART00000100737
|
acot17
|
acyl-CoA thioesterase 17 |
| chr3_-_24681404 | 1.02 |
ENSDART00000161612
|
BX569789.1
|
|
| chr6_-_27108844 | 1.00 |
ENSDART00000073883
|
dtymk
|
deoxythymidylate kinase (thymidylate kinase) |
| chr22_+_9918872 | 0.94 |
ENSDART00000177953
|
blf
|
bloody fingers |
| chr2_-_14798295 | 0.93 |
ENSDART00000143430
ENSDART00000145869 |
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr5_+_26121393 | 0.92 |
ENSDART00000002221
|
bco2l
|
beta-carotene 15, 15-dioxygenase 2, like |
| chr2_-_14793343 | 0.91 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr4_+_72235562 | 0.89 |
ENSDART00000168547
|
si:cabz01071912.2
|
si:cabz01071912.2 |
| chr13_-_33683889 | 0.89 |
ENSDART00000136820
ENSDART00000065435 |
cst3
|
cystatin C (amyloid angiopathy and cerebral hemorrhage) |
| chr15_-_18361475 | 0.87 |
ENSDART00000155866
|
zbtb16b
|
zinc finger and BTB domain containing 16b |
| chr11_+_14333441 | 0.85 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr23_+_19790962 | 0.82 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr14_-_9277152 | 0.82 |
ENSDART00000189048
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr9_+_15890558 | 0.81 |
ENSDART00000144032
|
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr18_+_20468157 | 0.79 |
ENSDART00000100665
ENSDART00000147867 ENSDART00000060302 ENSDART00000180370 |
ddb2
|
damage-specific DNA binding protein 2 |
| chr3_-_22228602 | 0.78 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr22_-_15602760 | 0.78 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr13_+_18321140 | 0.76 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr21_-_36619599 | 0.76 |
ENSDART00000065208
|
nop16
|
NOP16 nucleolar protein homolog (yeast) |
| chr1_-_999556 | 0.75 |
ENSDART00000170884
ENSDART00000172235 |
gart
|
phosphoribosylglycinamide formyltransferase |
| chr16_+_23960933 | 0.74 |
ENSDART00000146077
|
apoeb
|
apolipoprotein Eb |
| chr20_-_47188966 | 0.72 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr22_-_15602593 | 0.68 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr7_+_12835048 | 0.67 |
ENSDART00000016465
|
cx36.7
|
connexin 36.7 |
| chr7_-_30174882 | 0.67 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr19_+_17642356 | 0.67 |
ENSDART00000176431
|
CR382334.1
|
|
| chr11_-_20956309 | 0.67 |
ENSDART00000188659
|
CABZ01008739.1
|
|
| chr21_+_19858627 | 0.64 |
ENSDART00000147010
|
fybb
|
FYN binding protein b |
| chr21_-_2958422 | 0.64 |
ENSDART00000174091
|
zgc:194215
|
zgc:194215 |
| chr5_+_53482597 | 0.62 |
ENSDART00000180333
|
BX323994.1
|
|
| chr4_+_51347000 | 0.61 |
ENSDART00000161859
|
si:dkeyp-82b4.3
|
si:dkeyp-82b4.3 |
| chr16_-_39131666 | 0.61 |
ENSDART00000075517
|
gdf6a
|
growth differentiation factor 6a |
| chr12_+_27068525 | 0.61 |
ENSDART00000188634
|
srcap
|
Snf2-related CREBBP activator protein |
| chr13_+_15682803 | 0.60 |
ENSDART00000188063
|
CR931980.1
|
|
| chr6_+_32834760 | 0.59 |
ENSDART00000121562
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr14_+_6954579 | 0.57 |
ENSDART00000060998
|
nme5
|
NME/NM23 family member 5 |
| chr1_+_47178529 | 0.56 |
ENSDART00000158432
ENSDART00000074450 ENSDART00000137448 |
morc3b
|
MORC family CW-type zinc finger 3b |
| chr3_-_58644920 | 0.55 |
ENSDART00000155953
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr16_+_23961276 | 0.55 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr7_+_21917857 | 0.53 |
ENSDART00000088041
|
ufsp1
|
UFM1-specific peptidase 1 (non-functional) |
| chr16_-_13595027 | 0.52 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr3_-_32541033 | 0.52 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr16_+_27444098 | 0.51 |
ENSDART00000157690
|
invs
|
inversin |
| chr7_+_23943597 | 0.51 |
ENSDART00000157408
|
si:dkey-183c6.7
|
si:dkey-183c6.7 |
| chr14_+_11458044 | 0.51 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr4_-_8030583 | 0.50 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr3_-_32603191 | 0.50 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr19_-_25464291 | 0.50 |
ENSDART00000112915
|
umad1
|
UBAP1-MVB12-associated (UMA) domain containing 1 |
| chr14_+_11457500 | 0.49 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr13_+_21676235 | 0.49 |
ENSDART00000137804
ENSDART00000134950 ENSDART00000129653 |
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr6_-_49078263 | 0.49 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr15_-_29388012 | 0.49 |
ENSDART00000115032
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr21_-_31207522 | 0.49 |
ENSDART00000191637
|
zgc:152891
|
zgc:152891 |
| chr4_+_70556298 | 0.48 |
ENSDART00000170985
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr8_+_21159122 | 0.47 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr16_-_26727032 | 0.46 |
ENSDART00000177668
|
rnf41l
|
ring finger protein 41, like |
| chr3_+_31600593 | 0.46 |
ENSDART00000076640
ENSDART00000148189 |
ccdc43
|
coiled-coil domain containing 43 |
| chr6_-_24301324 | 0.45 |
ENSDART00000171401
|
BX927081.1
|
|
| chr9_+_21993092 | 0.45 |
ENSDART00000059693
|
crygm7
|
crystallin, gamma M7 |
| chr23_+_36616717 | 0.45 |
ENSDART00000042701
ENSDART00000192980 |
pip4k2ca
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma a |
| chr22_+_5478353 | 0.45 |
ENSDART00000160596
|
tppp
|
tubulin polymerization promoting protein |
| chr13_+_21677767 | 0.44 |
ENSDART00000165166
|
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr5_+_57328535 | 0.44 |
ENSDART00000149646
|
slc31a1
|
solute carrier family 31 (copper transporter), member 1 |
| chr15_-_29387446 | 0.43 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr9_-_28937880 | 0.43 |
ENSDART00000132878
|
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr6_-_35052145 | 0.43 |
ENSDART00000073970
ENSDART00000185790 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr3_+_34988670 | 0.43 |
ENSDART00000011319
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr20_+_25581627 | 0.43 |
ENSDART00000030229
|
cyp2p9
|
cytochrome P450, family 2, subfamily P, polypeptide 9 |
| chr5_-_24517768 | 0.42 |
ENSDART00000003957
|
tia1l
|
cytotoxic granule-associated RNA binding protein 1, like |
| chr14_+_28438947 | 0.42 |
ENSDART00000006489
|
acsl4a
|
acyl-CoA synthetase long chain family member 4a |
| chr24_+_15655233 | 0.42 |
ENSDART00000143160
|
fbxo15
|
F-box protein 15 |
| chr12_-_37941733 | 0.42 |
ENSDART00000130167
|
CABZ01046949.1
|
|
| chr23_-_26880623 | 0.41 |
ENSDART00000038491
|
adcy6b
|
adenylate cyclase 6b |
| chr8_-_20138054 | 0.41 |
ENSDART00000133141
ENSDART00000147634 ENSDART00000029939 |
rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr24_+_14527935 | 0.41 |
ENSDART00000134846
|
si:dkeyp-73g8.5
|
si:dkeyp-73g8.5 |
| chr8_+_46536893 | 0.40 |
ENSDART00000124023
|
pimr187
|
Pim proto-oncogene, serine/threonine kinase, related 187 |
| chr1_-_22851481 | 0.40 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr2_+_15069011 | 0.40 |
ENSDART00000145893
|
cnn3b
|
calponin 3, acidic b |
| chr23_-_20361971 | 0.39 |
ENSDART00000133373
|
si:rp71-17i16.5
|
si:rp71-17i16.5 |
| chr14_-_21064199 | 0.39 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr9_+_16449398 | 0.39 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr17_-_25563847 | 0.39 |
ENSDART00000040032
|
rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr16_+_48682309 | 0.38 |
ENSDART00000183957
|
tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr22_+_10163901 | 0.38 |
ENSDART00000190468
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr16_+_38394371 | 0.38 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr14_+_18785727 | 0.37 |
ENSDART00000184452
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
| chr17_-_27223965 | 0.37 |
ENSDART00000192577
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr21_-_21465111 | 0.36 |
ENSDART00000141487
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr19_-_42556086 | 0.35 |
ENSDART00000051731
|
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr25_+_16079913 | 0.35 |
ENSDART00000146350
|
far1
|
fatty acyl CoA reductase 1 |
| chr23_+_5524247 | 0.35 |
ENSDART00000189679
ENSDART00000083622 |
tead3a
|
TEA domain family member 3 a |
| chr9_-_39968820 | 0.34 |
ENSDART00000100311
|
IKZF2
|
si:zfos-1425h8.1 |
| chr14_-_24101897 | 0.34 |
ENSDART00000143695
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr9_+_6587364 | 0.34 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr7_+_57108823 | 0.33 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr2_-_37477654 | 0.33 |
ENSDART00000193921
|
dapk3
|
death-associated protein kinase 3 |
| chr8_-_52715911 | 0.33 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr9_+_24126223 | 0.33 |
ENSDART00000132045
|
pgap1
|
post-GPI attachment to proteins 1 |
| chr13_+_30804367 | 0.32 |
ENSDART00000053946
|
cxcl12a
|
chemokine (C-X-C motif) ligand 12a (stromal cell-derived factor 1) |
| chr2_-_37889111 | 0.32 |
ENSDART00000168939
ENSDART00000098529 |
mbl2
|
mannose binding lectin 2 |
| chr18_+_26750516 | 0.32 |
ENSDART00000110843
|
alpk3a
|
alpha-kinase 3a |
| chr11_-_24016761 | 0.32 |
ENSDART00000153601
ENSDART00000067817 ENSDART00000170531 |
chia.3
|
chitinase, acidic.3 |
| chr3_-_61375496 | 0.32 |
ENSDART00000165188
|
si:dkey-111k8.2
|
si:dkey-111k8.2 |
| chr9_+_9927516 | 0.31 |
ENSDART00000161089
ENSDART00000168559 |
ttf2
|
transcription termination factor, RNA polymerase II |
| chr15_-_28082310 | 0.31 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr2_+_394166 | 0.31 |
ENSDART00000155733
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr8_+_27555314 | 0.31 |
ENSDART00000135568
ENSDART00000016696 |
rhocb
|
ras homolog family member Cb |
| chr1_+_51191049 | 0.30 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr7_-_39751540 | 0.30 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr5_+_37087583 | 0.30 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr7_+_59649399 | 0.29 |
ENSDART00000123520
ENSDART00000040771 |
rpl34
|
ribosomal protein L34 |
| chr7_-_72382122 | 0.29 |
ENSDART00000191418
|
CU459012.3
|
|
| chr18_-_40481028 | 0.29 |
ENSDART00000134177
|
zgc:101040
|
zgc:101040 |
| chr2_+_19195841 | 0.29 |
ENSDART00000163137
ENSDART00000161095 |
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr4_+_31456734 | 0.29 |
ENSDART00000140176
|
znf996
|
zinc finger protein 996 |
| chr13_+_39297802 | 0.28 |
ENSDART00000133636
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr17_+_7595356 | 0.28 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr22_+_9522971 | 0.28 |
ENSDART00000110048
|
strip1
|
striatin interacting protein 1 |
| chr19_-_24443867 | 0.28 |
ENSDART00000163763
ENSDART00000043133 |
thbs3b
|
thrombospondin 3b |
| chr13_+_18545819 | 0.28 |
ENSDART00000101859
ENSDART00000110197 ENSDART00000136064 |
zgc:154058
|
zgc:154058 |
| chr25_-_27729046 | 0.28 |
ENSDART00000131437
|
zgc:153935
|
zgc:153935 |
| chr17_-_14451405 | 0.27 |
ENSDART00000146786
|
jkamp
|
jnk1/mapk8 associated membrane protein |
| chr8_-_31416403 | 0.26 |
ENSDART00000098912
|
znf131
|
zinc finger protein 131 |
| chr12_-_36260532 | 0.26 |
ENSDART00000022533
|
kcnj2a
|
potassium inwardly-rectifying channel, subfamily J, member 2a |
| chr17_+_25563979 | 0.26 |
ENSDART00000045615
ENSDART00000183162 |
qrsl1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr17_+_6382275 | 0.26 |
ENSDART00000056324
|
CR628323.2
|
|
| chr8_+_28469054 | 0.26 |
ENSDART00000062716
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr2_-_11648731 | 0.25 |
ENSDART00000080925
|
impad1
|
inositol monophosphatase domain containing 1 |
| chr2_+_9536021 | 0.25 |
ENSDART00000187357
|
CU929379.1
|
|
| chr3_+_43955864 | 0.25 |
ENSDART00000168267
|
BX571715.1
|
|
| chr23_-_25135046 | 0.25 |
ENSDART00000184844
ENSDART00000103989 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr13_-_27384697 | 0.25 |
ENSDART00000146230
|
kcnq5a
|
potassium voltage-gated channel, KQT-like subfamily, member 5a |
| chr10_+_39263827 | 0.25 |
ENSDART00000172509
|
foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr11_-_22372072 | 0.25 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr5_+_53009083 | 0.25 |
ENSDART00000157836
|
hint2
|
histidine triad nucleotide binding protein 2 |
| chr23_+_21414005 | 0.25 |
ENSDART00000144686
|
iffo2a
|
intermediate filament family orphan 2a |
| chr5_-_15264007 | 0.24 |
ENSDART00000180641
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr19_+_30990815 | 0.24 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr2_+_40294313 | 0.23 |
ENSDART00000037292
|
epha4b
|
eph receptor A4b |
| chr3_-_8130926 | 0.23 |
ENSDART00000189495
|
si:ch211-51i16.1
|
si:ch211-51i16.1 |
| chr6_+_40437987 | 0.23 |
ENSDART00000136487
|
ghrl
|
ghrelin/obestatin prepropeptide |
| chr23_+_28381260 | 0.23 |
ENSDART00000162722
|
zgc:153867
|
zgc:153867 |
| chr3_-_40976288 | 0.23 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr22_+_25184459 | 0.23 |
ENSDART00000105308
|
si:ch211-226h8.4
|
si:ch211-226h8.4 |
| chr19_+_10603405 | 0.22 |
ENSDART00000151135
|
si:dkey-211g8.8
|
si:dkey-211g8.8 |
| chr2_-_2642476 | 0.22 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr24_+_21174851 | 0.22 |
ENSDART00000154940
ENSDART00000155977 ENSDART00000122762 |
naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr23_+_12852655 | 0.22 |
ENSDART00000183015
|
smc1al
|
structural maintenance of chromosomes 1A, like |
| chr1_-_54706039 | 0.22 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr5_-_10239079 | 0.22 |
ENSDART00000132739
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr19_+_10831362 | 0.22 |
ENSDART00000053325
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr12_+_26632448 | 0.22 |
ENSDART00000185762
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr2_+_16191852 | 0.21 |
ENSDART00000152046
|
sh3glb1b
|
SH3-domain GRB2-like endophilin B1b |
| chr7_+_59212666 | 0.21 |
ENSDART00000172046
|
dok1b
|
docking protein 1b |
| chr23_+_22293682 | 0.21 |
ENSDART00000187304
|
CR354612.1
|
|
| chr11_-_3629201 | 0.20 |
ENSDART00000136577
ENSDART00000132121 |
itih3a
|
inter-alpha-trypsin inhibitor heavy chain 3a |
| chr15_+_38308421 | 0.19 |
ENSDART00000129941
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr17_+_50701748 | 0.19 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr14_+_22076596 | 0.19 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr3_-_29891456 | 0.18 |
ENSDART00000151677
ENSDART00000014021 |
slc25a39
|
solute carrier family 25, member 39 |
| chr17_-_48743076 | 0.18 |
ENSDART00000173117
|
kcnk17
|
potassium channel, subfamily K, member 17 |
| chr16_-_55028740 | 0.18 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr9_+_32428679 | 0.18 |
ENSDART00000187650
ENSDART00000190472 |
plcl1
|
phospholipase C like 1 |
| chr17_+_14784275 | 0.18 |
ENSDART00000005355
|
rtraf
|
RNA transcription, translation and transport factor |
| chr6_-_1587291 | 0.18 |
ENSDART00000067592
ENSDART00000178877 |
zgc:123305
|
zgc:123305 |
| chr16_-_28876479 | 0.18 |
ENSDART00000047862
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr10_-_17745345 | 0.18 |
ENSDART00000132690
ENSDART00000135376 |
si:dkey-200l5.4
|
si:dkey-200l5.4 |
| chr23_+_24989387 | 0.18 |
ENSDART00000172299
ENSDART00000145307 |
arhgap4a
|
Rho GTPase activating protein 4a |
| chr16_-_49846359 | 0.18 |
ENSDART00000184246
|
znf385d
|
zinc finger protein 385D |
| chr9_-_2572790 | 0.18 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr21_-_11632403 | 0.18 |
ENSDART00000171708
ENSDART00000138619 ENSDART00000136308 ENSDART00000144770 |
cast
|
calpastatin |
| chr14_-_25444774 | 0.17 |
ENSDART00000183448
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr3_-_16010968 | 0.17 |
ENSDART00000080672
|
mrps34
|
mitochondrial ribosomal protein S34 |
| chr5_+_57641554 | 0.17 |
ENSDART00000171309
ENSDART00000157992 ENSDART00000164742 |
cryabb
|
crystallin, alpha B, b |
| chr1_+_22851261 | 0.17 |
ENSDART00000193925
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr6_+_8630355 | 0.17 |
ENSDART00000161749
ENSDART00000193976 |
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr7_-_21928826 | 0.16 |
ENSDART00000088043
|
si:dkey-85k7.11
|
si:dkey-85k7.11 |
| chr12_+_30723778 | 0.16 |
ENSDART00000153291
|
slc35f3a
|
solute carrier family 35, member F3a |
| chr20_+_48543492 | 0.16 |
ENSDART00000175480
|
LO018254.1
|
|
| chr18_-_33093705 | 0.16 |
ENSDART00000059500
|
olfcd3
|
olfactory receptor C family, d3 |
| chr6_-_12135741 | 0.16 |
ENSDART00000155090
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr22_+_5176255 | 0.16 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr4_-_35900331 | 0.16 |
ENSDART00000164413
|
zgc:174653
|
zgc:174653 |
| chr24_+_26432541 | 0.16 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr13_-_9024004 | 0.16 |
ENSDART00000169564
|
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr3_-_39198113 | 0.15 |
ENSDART00000102690
|
RETSAT
|
zgc:154169 |
| chr22_+_12477996 | 0.15 |
ENSDART00000177704
|
CR847870.3
|
|
| chr22_+_34663328 | 0.15 |
ENSDART00000183912
|
si:ch73-304f21.1
|
si:ch73-304f21.1 |
| chr13_-_9895564 | 0.15 |
ENSDART00000169831
ENSDART00000142629 |
si:ch211-117n7.6
|
si:ch211-117n7.6 |
| chr22_-_11729350 | 0.15 |
ENSDART00000105813
|
krt222
|
keratin 222 |
| chr20_+_23960525 | 0.15 |
ENSDART00000042123
|
cx52.6
|
connexin 52.6 |
| chr3_-_26921475 | 0.14 |
ENSDART00000130281
|
ciita
|
class II, major histocompatibility complex, transactivator |
| chr19_-_12404590 | 0.14 |
ENSDART00000103703
|
ftr56
|
finTRIM family, member 56 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfixa+nfixb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.4 | 1.3 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.4 | 1.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 | 1.0 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.2 | 0.5 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 0.6 | GO:0042671 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.7 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.1 | 0.4 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.5 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 0.4 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.1 | 0.3 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.1 | 0.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.1 | 0.4 | GO:0019884 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.4 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.4 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.4 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.1 | 0.8 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 0.4 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.1 | 0.2 | GO:0071962 | establishment of mitotic sister chromatid cohesion(GO:0034087) mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.1 | 0.4 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.0 | 1.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.3 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 1.3 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.9 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.8 | GO:0009411 | response to UV(GO:0009411) |
| 0.0 | 0.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.2 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.2 | GO:0032095 | regulation of response to food(GO:0032095) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.7 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 1.1 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.4 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 0.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.4 | GO:0048515 | spermatid development(GO:0007286) spermatid differentiation(GO:0048515) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.5 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 1.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.4 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0098576 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) lumenal side of membrane(GO:0098576) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 2.9 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.1 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.0 | 1.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 1.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 0.5 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 0.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.9 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.4 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.7 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 0.4 | GO:0015440 | peptide antigen-transporting ATPase activity(GO:0015433) peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) |
| 0.1 | 0.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.2 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.1 | 0.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.5 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 1.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.1 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 2.3 | GO:0042802 | identical protein binding(GO:0042802) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |