Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
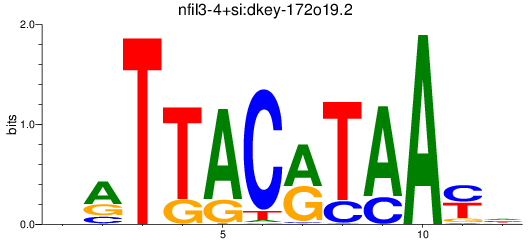
Results for nfil3-4+si:dkey-172o19.2
Z-value: 1.88
Transcription factors associated with nfil3-4+si:dkey-172o19.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_dkey-172o19.2
|
ENSDARG00000071398 | si_dkey-172o19.2 |
|
nfil3-4
|
ENSDARG00000092346 | nuclear factor, interleukin 3 regulated, member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:dkey-172o19.2 | dr11_v1_chr22_+_20546612_20546612 | 0.21 | 3.8e-01 | Click! |
| nfil3-4 | dr11_v1_chr22_+_20560041_20560041 | 0.12 | 6.4e-01 | Click! |
Activity profile of nfil3-4+si:dkey-172o19.2 motif
Sorted Z-values of nfil3-4+si:dkey-172o19.2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_345503 | 4.54 |
ENSDART00000168901
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
| chr20_-_25533739 | 3.57 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr5_+_45677781 | 3.45 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr11_-_29996344 | 3.39 |
ENSDART00000003712
ENSDART00000126110 |
ace2
|
angiotensin I converting enzyme 2 |
| chr11_-_8167799 | 3.29 |
ENSDART00000133574
ENSDART00000024046 ENSDART00000146940 |
uox
|
urate oxidase |
| chr12_+_46960579 | 3.21 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr9_-_48736388 | 3.07 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr17_+_27456804 | 2.81 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr25_-_29415369 | 2.80 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr6_-_16406210 | 2.78 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr10_+_38526496 | 2.76 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr21_-_43550120 | 2.64 |
ENSDART00000151627
|
si:ch73-362m14.2
|
si:ch73-362m14.2 |
| chr25_-_8602437 | 2.63 |
ENSDART00000171200
|
rhcgb
|
Rh family, C glycoprotein b |
| chr20_-_44576949 | 2.59 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr25_+_34984333 | 2.48 |
ENSDART00000154760
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr19_+_5674907 | 2.44 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr5_+_51079504 | 2.42 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr20_-_25522911 | 2.36 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr8_-_32497815 | 2.35 |
ENSDART00000122359
|
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr13_+_33688474 | 2.28 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr2_-_32643738 | 2.20 |
ENSDART00000112452
|
si:dkeyp-73d8.9
|
si:dkeyp-73d8.9 |
| chr18_+_41232719 | 2.13 |
ENSDART00000138552
ENSDART00000145863 |
trip12
|
thyroid hormone receptor interactor 12 |
| chr19_+_9174166 | 2.07 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr17_+_15845765 | 2.07 |
ENSDART00000130881
ENSDART00000074936 |
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr3_+_21225750 | 2.06 |
ENSDART00000139213
|
zgc:153968
|
zgc:153968 |
| chr12_-_32183164 | 2.05 |
ENSDART00000191269
ENSDART00000173549 |
SMIM36
|
si:ch73-256g18.2 |
| chr1_-_9231952 | 2.03 |
ENSDART00000166515
|
si:dkeyp-57d7.4
|
si:dkeyp-57d7.4 |
| chr25_+_4760489 | 2.03 |
ENSDART00000167399
|
FP245455.1
|
|
| chr5_+_4332220 | 2.03 |
ENSDART00000051699
|
sat1a.1
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 1 |
| chr9_-_44953349 | 2.00 |
ENSDART00000135156
|
vil1
|
villin 1 |
| chr7_+_30371893 | 1.95 |
ENSDART00000075513
|
aqp9b
|
aquaporin 9b |
| chr13_-_293250 | 1.88 |
ENSDART00000138581
|
chs1
|
chitin synthase 1 |
| chr8_-_39739627 | 1.86 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr5_-_2112030 | 1.86 |
ENSDART00000091932
|
gusb
|
glucuronidase, beta |
| chr23_+_33963619 | 1.84 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr5_-_57655092 | 1.84 |
ENSDART00000074290
|
mia
|
melanoma inhibitory activity |
| chr5_+_26121393 | 1.83 |
ENSDART00000002221
|
bco2l
|
beta-carotene 15, 15-dioxygenase 2, like |
| chr20_-_1151265 | 1.83 |
ENSDART00000012376
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr24_-_40009446 | 1.83 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr21_+_43178831 | 1.82 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr6_+_36381709 | 1.80 |
ENSDART00000004727
|
rhcgl1
|
Rh family, C glycoprotein, like 1 |
| chr16_-_4640539 | 1.77 |
ENSDART00000076955
ENSDART00000131949 |
cyp4t8
|
cytochrome P450, family 4, subfamily T, polypeptide 8 |
| chr8_-_46581448 | 1.75 |
ENSDART00000014240
|
sult1st3
|
sulfotransferase family 1, cytosolic sulfotransferase 3 |
| chr3_-_50443607 | 1.73 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr8_-_10045043 | 1.72 |
ENSDART00000081917
|
si:dkey-8e10.3
|
si:dkey-8e10.3 |
| chr22_+_15315655 | 1.71 |
ENSDART00000141249
|
sult3st3
|
sulfotransferase family 3, cytosolic sulfotransferase 3 |
| chr24_-_17023392 | 1.71 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr4_+_5531583 | 1.70 |
ENSDART00000137500
ENSDART00000042080 |
si:dkey-14d8.6
|
si:dkey-14d8.6 |
| chr20_+_1121458 | 1.69 |
ENSDART00000064472
|
pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr4_-_9549693 | 1.68 |
ENSDART00000160242
|
FQ377934.1
|
|
| chr13_-_25750910 | 1.68 |
ENSDART00000111567
|
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr15_-_34418525 | 1.67 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr5_+_2815021 | 1.67 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr20_-_40755614 | 1.66 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr4_+_18804317 | 1.64 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr4_+_5537101 | 1.62 |
ENSDART00000008692
|
si:dkey-14d8.7
|
si:dkey-14d8.7 |
| chr10_+_38417512 | 1.62 |
ENSDART00000112457
|
samsn1b
|
SAM domain, SH3 domain and nuclear localisation signals 1b |
| chr8_-_8346023 | 1.62 |
ENSDART00000164731
ENSDART00000188259 |
tsr2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr9_-_21238159 | 1.61 |
ENSDART00000146764
ENSDART00000102143 |
cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr4_-_22671469 | 1.61 |
ENSDART00000050753
|
cd36
|
CD36 molecule (thrombospondin receptor) |
| chr22_+_15336752 | 1.59 |
ENSDART00000139070
|
sult3st2
|
sulfotransferase family 3, cytosolic sulfotransferase 2 |
| chr5_-_25576462 | 1.58 |
ENSDART00000165147
|
si:dkey-229d2.4
|
si:dkey-229d2.4 |
| chr11_+_6295370 | 1.58 |
ENSDART00000139882
|
ranbp3a
|
RAN binding protein 3a |
| chr2_-_55337585 | 1.55 |
ENSDART00000177924
|
tpm4b
|
tropomyosin 4b |
| chr19_-_20093341 | 1.53 |
ENSDART00000129917
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr20_-_34801181 | 1.53 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr24_-_4765740 | 1.53 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr5_-_69212184 | 1.52 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr7_-_6604623 | 1.48 |
ENSDART00000172874
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr3_-_53465223 | 1.48 |
ENSDART00000057123
ENSDART00000125515 ENSDART00000143096 |
nr5a5
|
nuclear receptor subfamily 5, group A, member 5 |
| chr15_+_37559570 | 1.46 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr6_-_39649504 | 1.44 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr19_-_657439 | 1.43 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr7_+_39385540 | 1.42 |
ENSDART00000139240
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr7_-_26457208 | 1.42 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr4_-_77432218 | 1.41 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr1_-_54947592 | 1.41 |
ENSDART00000129710
|
crtac1a
|
cartilage acidic protein 1a |
| chr23_-_30785382 | 1.41 |
ENSDART00000136156
ENSDART00000131285 |
myt1a
|
myelin transcription factor 1a |
| chr16_-_11798994 | 1.41 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr8_-_37263524 | 1.41 |
ENSDART00000061327
|
rh50
|
Rh50-like protein |
| chr18_+_36037223 | 1.40 |
ENSDART00000144410
|
tmem91
|
transmembrane protein 91 |
| chr2_+_10134345 | 1.38 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr2_+_24177190 | 1.38 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr7_+_15659280 | 1.36 |
ENSDART00000173414
|
mef2ab
|
myocyte enhancer factor 2ab |
| chr9_-_44953664 | 1.35 |
ENSDART00000188558
ENSDART00000185210 |
vil1
|
villin 1 |
| chr8_-_32497581 | 1.35 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr19_+_7735157 | 1.34 |
ENSDART00000186717
|
tuft1b
|
tuftelin 1b |
| chr23_+_5108374 | 1.33 |
ENSDART00000114263
|
zgc:194242
|
zgc:194242 |
| chr2_+_7192966 | 1.32 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr3_+_16612574 | 1.31 |
ENSDART00000104481
|
slc17a7a
|
solute carrier family 17 (vesicular glutamate transporter), member 7a |
| chr21_-_43949208 | 1.29 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr17_-_5610514 | 1.29 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr9_-_46399611 | 1.29 |
ENSDART00000164914
ENSDART00000145931 |
lct
si:dkey-79p17.3
|
lactase si:dkey-79p17.3 |
| chr5_-_67878064 | 1.29 |
ENSDART00000111203
|
tagln3a
|
transgelin 3a |
| chr8_+_44714336 | 1.29 |
ENSDART00000145801
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr9_-_40846447 | 1.28 |
ENSDART00000143384
|
si:dkey-95p16.1
|
si:dkey-95p16.1 |
| chr22_-_10110959 | 1.28 |
ENSDART00000031005
ENSDART00000147580 |
gls2b
|
glutaminase 2b (liver, mitochondrial) |
| chr20_-_52939501 | 1.27 |
ENSDART00000166508
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr2_+_35603637 | 1.27 |
ENSDART00000147278
|
plk3
|
polo-like kinase 3 (Drosophila) |
| chr2_-_56635744 | 1.27 |
ENSDART00000167790
ENSDART00000168160 |
pip5k1cb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b |
| chr17_+_10566490 | 1.26 |
ENSDART00000144408
ENSDART00000137469 |
mgaa
|
MGA, MAX dimerization protein a |
| chr24_-_10897511 | 1.26 |
ENSDART00000145593
ENSDART00000102484 ENSDART00000066784 |
fam49bb
|
family with sequence similarity 49, member Bb |
| chr16_-_46587938 | 1.24 |
ENSDART00000181433
|
BX323793.1
|
|
| chr15_-_34408777 | 1.24 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr1_+_47486104 | 1.24 |
ENSDART00000114746
|
lrrc58a
|
leucine rich repeat containing 58a |
| chr16_-_13818061 | 1.23 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr21_+_31151197 | 1.23 |
ENSDART00000137728
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr3_+_15296824 | 1.22 |
ENSDART00000043801
|
cabp5b
|
calcium binding protein 5b |
| chr21_-_5856050 | 1.22 |
ENSDART00000115367
|
CABZ01071020.1
|
|
| chr2_-_51772438 | 1.22 |
ENSDART00000170241
|
BX908782.2
|
Danio rerio three-finger protein 5 (LOC100003647), mRNA. |
| chr22_+_27090136 | 1.20 |
ENSDART00000136770
|
si:dkey-246e1.3
|
si:dkey-246e1.3 |
| chr18_+_13162728 | 1.20 |
ENSDART00000101472
|
tat
|
tyrosine aminotransferase |
| chr8_+_23703680 | 1.19 |
ENSDART00000141099
ENSDART00000135394 |
ppardb
|
peroxisome proliferator-activated receptor delta b |
| chr4_-_74998614 | 1.19 |
ENSDART00000162529
|
zgc:172139
|
zgc:172139 |
| chr21_+_25801345 | 1.19 |
ENSDART00000035062
|
nf2b
|
neurofibromin 2b (merlin) |
| chr21_-_22910520 | 1.19 |
ENSDART00000065567
ENSDART00000191792 |
guca1d
|
guanylate cyclase activator 1d |
| chr3_+_49043917 | 1.18 |
ENSDART00000158212
|
zgc:92161
|
zgc:92161 |
| chr10_+_36650222 | 1.17 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr16_-_16212615 | 1.16 |
ENSDART00000059905
|
upp1
|
uridine phosphorylase 1 |
| chr13_+_1542493 | 1.16 |
ENSDART00000181968
|
CABZ01044281.1
|
|
| chr15_-_43995028 | 1.15 |
ENSDART00000172485
ENSDART00000186320 |
nlrc3l
|
NLR family, CARD domain containing 3-like |
| chr10_-_24371312 | 1.15 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr21_-_39931285 | 1.15 |
ENSDART00000180010
ENSDART00000024407 |
tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr2_-_11512819 | 1.14 |
ENSDART00000142013
|
penka
|
proenkephalin a |
| chr16_-_33104944 | 1.13 |
ENSDART00000151943
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr23_-_18030399 | 1.12 |
ENSDART00000136967
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr6_-_43449013 | 1.10 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr1_-_20928772 | 1.10 |
ENSDART00000078277
|
msmo1
|
methylsterol monooxygenase 1 |
| chr23_-_4925131 | 1.10 |
ENSDART00000138805
|
taz
|
tafazzin |
| chr25_+_24250247 | 1.10 |
ENSDART00000064646
|
tmem86a
|
transmembrane protein 86A |
| chr22_+_1556948 | 1.09 |
ENSDART00000159050
|
si:ch211-255f4.8
|
si:ch211-255f4.8 |
| chr9_+_33154841 | 1.09 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr23_-_33361425 | 1.09 |
ENSDART00000031638
|
slc48a1a
|
solute carrier family 48 (heme transporter), member 1a |
| chr6_-_38818582 | 1.08 |
ENSDART00000149833
|
cnga3a
|
cyclic nucleotide gated channel alpha 3a |
| chr17_-_33416020 | 1.08 |
ENSDART00000140149
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr11_+_6819050 | 1.07 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr23_+_19590006 | 1.07 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr15_-_26636826 | 1.06 |
ENSDART00000087632
|
slc47a4
|
solute carrier family 47 (multidrug and toxin extrusion), member 4 |
| chr8_+_42917515 | 1.06 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr5_-_8483457 | 1.05 |
ENSDART00000171886
ENSDART00000159477 ENSDART00000157423 |
lifra
|
leukemia inhibitory factor receptor alpha a |
| chr6_-_49873020 | 1.05 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr7_-_12909352 | 1.05 |
ENSDART00000172901
|
sh3gl3a
|
SH3-domain GRB2-like 3a |
| chr2_-_55779927 | 1.05 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr11_+_25278772 | 1.05 |
ENSDART00000188630
|
cyldb
|
cylindromatosis (turban tumor syndrome), b |
| chr19_-_25005609 | 1.04 |
ENSDART00000151129
|
xkr8.2
|
XK, Kell blood group complex subunit-related family, member 8, tandem duplicate 2 |
| chr21_+_45841731 | 1.04 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr12_+_33396489 | 1.04 |
ENSDART00000149960
|
fasn
|
fatty acid synthase |
| chr4_+_5132951 | 1.03 |
ENSDART00000103279
|
ccnd2b
|
cyclin D2, b |
| chr23_+_19590598 | 1.03 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr3_+_7763114 | 1.02 |
ENSDART00000057434
|
hook2
|
hook microtubule-tethering protein 2 |
| chr11_+_25504215 | 1.02 |
ENSDART00000154213
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr2_-_20890429 | 1.02 |
ENSDART00000149989
|
pdca
|
phosducin a |
| chr11_-_44931962 | 1.01 |
ENSDART00000170345
|
pfklb
|
phosphofructokinase, liver b |
| chr20_+_38201644 | 1.01 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr20_+_25563105 | 1.01 |
ENSDART00000063100
|
cyp2p6
|
cytochrome P450, family 2, subfamily P, polypeptide 6 |
| chr13_-_301309 | 1.01 |
ENSDART00000131747
|
chs1
|
chitin synthase 1 |
| chr15_+_45640906 | 1.01 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr23_+_4022620 | 1.00 |
ENSDART00000055099
|
b4galt5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr7_+_30392613 | 1.00 |
ENSDART00000075508
|
lipca
|
lipase, hepatic a |
| chr20_-_51727860 | 1.00 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr14_+_4151379 | 0.99 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr14_-_5642371 | 0.99 |
ENSDART00000183859
ENSDART00000054876 |
npm1b
|
nucleophosmin 1b |
| chr18_+_13164325 | 0.98 |
ENSDART00000189057
|
tat
|
tyrosine aminotransferase |
| chr7_-_41693004 | 0.98 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr22_-_24297510 | 0.97 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr11_-_34151487 | 0.97 |
ENSDART00000173039
|
atp13a3
|
ATPase 13A3 |
| chr2_-_39017838 | 0.96 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr5_-_68244564 | 0.96 |
ENSDART00000169350
|
CABZ01083944.1
|
|
| chr17_+_8184649 | 0.96 |
ENSDART00000091818
|
tulp4b
|
tubby like protein 4b |
| chr7_+_6915609 | 0.95 |
ENSDART00000159213
|
ccs
|
copper chaperone for superoxide dismutase |
| chr12_-_32066469 | 0.95 |
ENSDART00000140685
ENSDART00000062185 |
rab40b
|
RAB40B, member RAS oncogene family |
| chr17_-_2690083 | 0.94 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr15_-_5016039 | 0.94 |
ENSDART00000156458
|
defbl3
|
defensin, beta-like 3 |
| chr3_+_41731527 | 0.93 |
ENSDART00000049007
ENSDART00000187866 |
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr23_-_44786844 | 0.93 |
ENSDART00000148669
|
si:ch73-269m23.5
|
si:ch73-269m23.5 |
| chr16_-_17175731 | 0.93 |
ENSDART00000183057
|
opn9
|
opsin 9 |
| chr15_+_45643787 | 0.93 |
ENSDART00000055995
ENSDART00000157750 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr18_+_507835 | 0.92 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr11_+_39107131 | 0.92 |
ENSDART00000105140
|
zgc:112255
|
zgc:112255 |
| chr5_-_55848358 | 0.92 |
ENSDART00000130891
|
camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr17_-_15274648 | 0.91 |
ENSDART00000141257
|
ros1
|
c-ros oncogene 1 , receptor tyrosine kinase |
| chr14_-_12071447 | 0.90 |
ENSDART00000166116
|
tmsb1
|
thymosin beta 1 |
| chr21_+_25221940 | 0.90 |
ENSDART00000108972
|
sycn.1
|
syncollin, tandem duplicate 1 |
| chr11_+_43401592 | 0.90 |
ENSDART00000112468
|
vipb
|
vasoactive intestinal peptide b |
| chr20_-_45661049 | 0.90 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr2_+_35240764 | 0.90 |
ENSDART00000015827
|
tnr
|
tenascin R (restrictin, janusin) |
| chr4_+_73651452 | 0.89 |
ENSDART00000174164
|
znf989
|
zinc finger protein 989 |
| chr16_+_39146696 | 0.89 |
ENSDART00000121756
ENSDART00000084381 |
sybu
|
syntabulin (syntaxin-interacting) |
| chr18_+_910992 | 0.89 |
ENSDART00000161206
ENSDART00000167229 |
pkma
|
pyruvate kinase M1/2a |
| chr6_+_8314451 | 0.89 |
ENSDART00000147793
ENSDART00000183688 |
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr12_+_3571770 | 0.88 |
ENSDART00000164707
ENSDART00000189819 |
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr3_-_42981739 | 0.88 |
ENSDART00000167844
|
mafk
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr9_+_42095220 | 0.88 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr13_-_20540790 | 0.88 |
ENSDART00000131467
|
si:ch1073-126c3.2
|
si:ch1073-126c3.2 |
| chr8_-_29851706 | 0.88 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr14_+_16345003 | 0.88 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr15_-_1885247 | 0.88 |
ENSDART00000149703
|
porb
|
P450 (cytochrome) oxidoreductase b |
| chr18_+_21061216 | 0.88 |
ENSDART00000141739
|
fam169b
|
family with sequence similarity 169, member B |
| chr5_+_1278092 | 0.88 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr24_-_38079261 | 0.87 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfil3-4+si:dkey-172o19.2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0019628 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.8 | 3.4 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.8 | 4.5 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.7 | 5.8 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.6 | 3.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.6 | 1.8 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.6 | 2.9 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.6 | 2.9 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.5 | 2.0 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.5 | 1.8 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.4 | 1.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.4 | 1.9 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.4 | 1.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.4 | 1.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.4 | 2.1 | GO:0044819 | mitotic G1/S transition checkpoint(GO:0044819) |
| 0.4 | 3.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.4 | 2.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.4 | 1.1 | GO:0044320 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.3 | 1.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.3 | 0.8 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.3 | 0.8 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.3 | 1.9 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.2 | 2.9 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.2 | 4.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.2 | 2.7 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.2 | 0.9 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.2 | 1.2 | GO:0014005 | microglia development(GO:0014005) |
| 0.2 | 0.7 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 0.7 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.2 | 0.9 | GO:0048241 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.2 | 0.9 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 0.7 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.2 | 0.9 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 1.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 1.9 | GO:0033273 | response to vitamin(GO:0033273) |
| 0.2 | 0.6 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.2 | 1.5 | GO:0036268 | swimming(GO:0036268) |
| 0.2 | 1.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 0.6 | GO:2000376 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.2 | 0.8 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.2 | 0.2 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.2 | 1.6 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 0.6 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.2 | 0.8 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.2 | 2.5 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.2 | 0.6 | GO:0010893 | positive regulation of steroid biosynthetic process(GO:0010893) |
| 0.2 | 0.9 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 1.3 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.2 | 0.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.2 | 5.1 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 3.0 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.2 | 0.5 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.2 | 1.7 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 0.8 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 1.8 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 1.9 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.2 | 1.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 0.6 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.2 | 0.8 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.2 | 1.3 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.2 | 7.7 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.2 | 0.8 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.2 | 1.4 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.2 | 0.6 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 1.5 | GO:0010138 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.2 | 0.8 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.4 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 2.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 0.9 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.6 | GO:0019477 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 1.1 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 3.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 1.7 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 0.9 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.5 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.8 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 3.4 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 1.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 4.7 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 0.6 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.2 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.1 | 1.1 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.9 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.1 | 1.1 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 1.0 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 0.5 | GO:0030328 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.1 | 0.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 2.6 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 1.1 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.6 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.7 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 1.6 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 0.7 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.5 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.9 | GO:0046471 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.1 | 0.3 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 0.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.9 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.0 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 2.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 1.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 1.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.2 | GO:0032979 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.3 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 3.4 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 0.5 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 0.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.7 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.1 | 0.3 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 0.4 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 0.3 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.7 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.5 | GO:0042493 | response to drug(GO:0042493) |
| 0.1 | 0.5 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 1.1 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 0.4 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 0.9 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.4 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.3 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 1.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.3 | GO:0032648 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 2.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) single-organism membrane invagination(GO:1902534) |
| 0.0 | 2.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.4 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.4 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.2 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.4 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.6 | GO:0006000 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.4 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.4 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.9 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.7 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.8 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 1.6 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.5 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.2 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.5 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 1.0 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 1.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.5 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.9 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.9 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.9 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.5 | GO:0072028 | nephron morphogenesis(GO:0072028) |
| 0.0 | 2.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.4 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.2 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.8 | GO:0007286 | spermatid development(GO:0007286) spermatid differentiation(GO:0048515) |
| 0.0 | 0.2 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.7 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.4 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.8 | GO:0000302 | response to reactive oxygen species(GO:0000302) |
| 0.0 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.3 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.2 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.8 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.5 | GO:0044275 | cellular carbohydrate catabolic process(GO:0044275) |
| 0.0 | 0.1 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 1.5 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 0.6 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 0.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.5 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 1.4 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 1.0 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.4 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 1.6 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.0 | 0.9 | GO:0009583 | phototransduction(GO:0007602) detection of light stimulus(GO:0009583) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0030816 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.8 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.0 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.5 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.5 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.1 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.4 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) organic acid transmembrane transport(GO:1903825) carboxylic acid transmembrane transport(GO:1905039) |
| 0.0 | 0.6 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:0019075 | virion assembly(GO:0019068) virus maturation(GO:0019075) receptor catabolic process(GO:0032801) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.0 | 2.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.4 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.6 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.5 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.1 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.4 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 1.0 | GO:0043434 | response to peptide hormone(GO:0043434) |
| 0.0 | 0.4 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.2 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.3 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.3 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.7 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.1 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 1.0 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.0 | 0.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.0 | 0.1 | GO:0090316 | positive regulation of intracellular protein transport(GO:0090316) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.6 | 4.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.4 | 1.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.4 | 2.9 | GO:0030428 | cell septum(GO:0030428) |
| 0.3 | 0.8 | GO:1990745 | EARP complex(GO:1990745) |
| 0.3 | 0.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 1.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 0.9 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.2 | 0.9 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 1.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 5.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.0 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 1.6 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 4.0 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.1 | 2.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 0.4 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.5 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.6 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.2 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) |
| 0.1 | 0.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 1.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.2 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.2 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 6.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.8 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.8 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 1.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.4 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.3 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 1.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.0 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 0.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.7 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 3.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.7 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 5.2 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 10.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 2.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.7 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 0.6 | GO:0098803 | respiratory chain complex(GO:0098803) |
| 0.0 | 2.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.2 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.0 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 1.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 1.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.6 | 1.8 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.5 | 1.9 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.5 | 2.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.5 | 1.4 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.5 | 2.8 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.4 | 3.4 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.4 | 1.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.4 | 1.7 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.4 | 1.6 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.4 | 1.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.4 | 1.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 3.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.4 | 3.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.4 | 1.5 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.4 | 2.9 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.3 | 4.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.3 | 1.0 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.3 | 1.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.3 | 1.3 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.3 | 2.9 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.3 | 2.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.3 | 1.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.3 | 2.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.3 | 1.6 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.3 | 1.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 1.0 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.2 | 1.0 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 0.7 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.2 | 2.1 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.2 | 5.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 1.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.2 | 0.9 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.2 | 0.9 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.2 | 0.6 | GO:0004788 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.2 | 0.6 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.2 | 1.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 0.6 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.2 | 1.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 0.6 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.2 | 1.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 0.6 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 5.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 1.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 1.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 0.9 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 1.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 0.7 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.2 | 1.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 5.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 0.8 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.2 | 1.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 1.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 1.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 1.0 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.1 | 0.9 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 2.0 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.7 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 0.4 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 1.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 1.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.8 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 5.2 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 8.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 1.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.5 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 1.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.3 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 1.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.6 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 0.4 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 0.5 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.1 | 0.5 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.1 | 1.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 1.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.6 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 0.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 1.8 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.5 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.5 | GO:0015250 | urea transmembrane transporter activity(GO:0015204) water channel activity(GO:0015250) |
| 0.1 | 0.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.2 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.1 | 0.5 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 1.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.9 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.6 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 3.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.5 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 2.1 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 1.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.5 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 2.0 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.5 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.3 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 0.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.4 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.3 | GO:0046625 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.0 | 0.6 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 3.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.1 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.0 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0008443 | 6-phosphofructo-2-kinase activity(GO:0003873) phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 2.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.9 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.3 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.4 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 1.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.8 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 1.5 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.5 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 2.4 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 3.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0072570 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.2 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.3 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.9 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.8 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.6 | GO:0017171 | serine-type peptidase activity(GO:0008236) serine hydrolase activity(GO:0017171) |
| 0.0 | 0.1 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.7 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 3.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.3 | 3.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 1.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 1.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 1.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 3.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.4 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 1.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.5 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.1 | 1.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.6 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 1.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 2.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 2.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.6 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.8 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 3.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.8 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.7 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |