Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
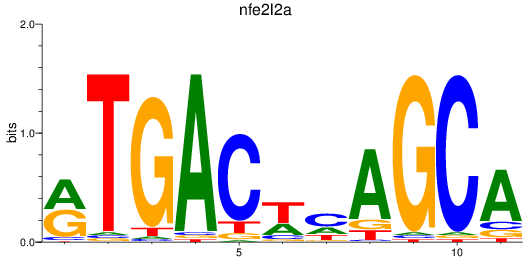
Results for nfe2l2a
Z-value: 0.88
Transcription factors associated with nfe2l2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfe2l2a
|
ENSDARG00000042824 | nuclear factor, erythroid 2-like 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfe2l2a | dr11_v1_chr9_+_1654284_1654284 | 0.80 | 3.3e-05 | Click! |
Activity profile of nfe2l2a motif
Sorted Z-values of nfe2l2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_67471375 | 4.66 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr5_-_30615901 | 4.66 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr10_-_7974155 | 3.13 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr13_+_28903959 | 2.79 |
ENSDART00000166435
|
si:ch73-28h20.1
|
si:ch73-28h20.1 |
| chr19_+_8506178 | 2.72 |
ENSDART00000189689
|
s100a10a
|
S100 calcium binding protein A10a |
| chr9_-_22076368 | 2.62 |
ENSDART00000128486
|
crygm2a
|
crystallin, gamma M2a |
| chr6_-_35446110 | 2.55 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr5_-_40297003 | 2.35 |
ENSDART00000097526
|
hspb3
|
heat shock protein, alpha-crystallin-related, b3 |
| chr11_+_24002503 | 2.29 |
ENSDART00000164702
|
chia.2
|
chitinase, acidic.2 |
| chr15_+_37559570 | 2.13 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr14_+_30279391 | 2.08 |
ENSDART00000172794
|
fgl1
|
fibrinogen-like 1 |
| chr2_+_9822319 | 2.08 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr20_-_39271844 | 2.02 |
ENSDART00000192708
|
clu
|
clusterin |
| chr6_+_21395051 | 1.99 |
ENSDART00000017774
|
cacng5a
|
calcium channel, voltage-dependent, gamma subunit 5a |
| chr23_+_19558574 | 1.84 |
ENSDART00000137811
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr24_-_29868151 | 1.83 |
ENSDART00000184802
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr18_+_1703984 | 1.83 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr3_-_21280373 | 1.78 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr1_+_6640437 | 1.75 |
ENSDART00000147638
|
si:ch211-93g23.2
|
si:ch211-93g23.2 |
| chr24_+_25259154 | 1.74 |
ENSDART00000171125
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr7_-_39552314 | 1.67 |
ENSDART00000134174
|
slc22a18
|
solute carrier family 22, member 18 |
| chr10_+_29963518 | 1.67 |
ENSDART00000011317
ENSDART00000099964 ENSDART00000182990 ENSDART00000113912 |
ntm
|
neurotrimin |
| chr23_-_18286822 | 1.64 |
ENSDART00000136672
|
fam19a1a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1a |
| chr13_-_21688176 | 1.62 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr22_-_16416882 | 1.61 |
ENSDART00000062749
|
cts12
|
cathepsin 12 |
| chr23_-_29376859 | 1.61 |
ENSDART00000146411
|
sst6
|
somatostatin 6 |
| chr17_+_27176243 | 1.58 |
ENSDART00000162527
|
si:ch211-160f23.7
|
si:ch211-160f23.7 |
| chr8_-_22660678 | 1.53 |
ENSDART00000181258
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr11_-_24681292 | 1.52 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr9_-_33749556 | 1.48 |
ENSDART00000149383
|
ndp
|
Norrie disease (pseudoglioma) |
| chr16_-_17162485 | 1.43 |
ENSDART00000123011
|
iffo1b
|
intermediate filament family orphan 1b |
| chr11_+_45300669 | 1.43 |
ENSDART00000172238
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr19_-_7450796 | 1.41 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr19_-_6385594 | 1.41 |
ENSDART00000104950
|
atp1a3a
|
ATPase Na+/K+ transporting subunit alpha 3a |
| chr8_+_24861264 | 1.41 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr2_+_9821757 | 1.40 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr8_-_34051548 | 1.40 |
ENSDART00000105204
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr7_-_39551913 | 1.39 |
ENSDART00000173498
ENSDART00000143040 |
slc22a18
|
solute carrier family 22, member 18 |
| chr19_+_48176745 | 1.37 |
ENSDART00000164963
|
prdm1b
|
PR domain containing 1b, with ZNF domain |
| chr14_-_48588422 | 1.33 |
ENSDART00000161147
|
si:ch211-154c21.1
|
si:ch211-154c21.1 |
| chr24_+_25258904 | 1.27 |
ENSDART00000155714
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr1_+_41690402 | 1.27 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr18_-_5692292 | 1.27 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr16_+_25259313 | 1.27 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr10_+_38775408 | 1.23 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr20_-_48604199 | 1.22 |
ENSDART00000161762
ENSDART00000170894 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr20_+_18661624 | 1.21 |
ENSDART00000152136
ENSDART00000126959 |
tnfaip2a
|
tumor necrosis factor, alpha-induced protein 2a |
| chr24_+_24923166 | 1.21 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr15_+_25438714 | 1.21 |
ENSDART00000154164
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr18_-_46763170 | 1.19 |
ENSDART00000171880
|
dner
|
delta/notch-like EGF repeat containing |
| chr11_-_9948487 | 1.19 |
ENSDART00000189677
ENSDART00000113171 |
nlgn1
|
neuroligin 1 |
| chr17_-_25737452 | 1.18 |
ENSDART00000152021
|
si:ch211-214p16.3
|
si:ch211-214p16.3 |
| chr1_-_40227166 | 1.17 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr7_-_49892991 | 1.15 |
ENSDART00000126240
|
cd44a
|
CD44 molecule (Indian blood group) a |
| chr9_+_11202552 | 1.13 |
ENSDART00000151931
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr7_-_15413019 | 1.08 |
ENSDART00000172147
|
si:ch211-106j24.1
|
si:ch211-106j24.1 |
| chr8_-_34052019 | 1.07 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr4_+_13696537 | 1.06 |
ENSDART00000109195
ENSDART00000122041 ENSDART00000192554 |
nrcama
|
neuronal cell adhesion molecule a |
| chr16_-_27640995 | 1.04 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr9_-_23253870 | 1.04 |
ENSDART00000143657
ENSDART00000169911 |
acmsd
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr4_-_16334362 | 1.04 |
ENSDART00000101461
|
epyc
|
epiphycan |
| chr10_+_26747755 | 1.02 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr22_-_31059670 | 1.02 |
ENSDART00000022445
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr18_-_19405616 | 0.99 |
ENSDART00000191290
ENSDART00000090855 |
megf11
|
multiple EGF-like-domains 11 |
| chr17_+_28533102 | 0.95 |
ENSDART00000156218
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr17_-_29902187 | 0.94 |
ENSDART00000009104
|
esrrb
|
estrogen-related receptor beta |
| chr7_-_52842605 | 0.88 |
ENSDART00000083002
|
map1aa
|
microtubule-associated protein 1Aa |
| chr22_-_13857729 | 0.88 |
ENSDART00000177971
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr18_+_26829086 | 0.88 |
ENSDART00000098356
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr19_-_32600823 | 0.87 |
ENSDART00000134149
ENSDART00000187858 |
zgc:91944
|
zgc:91944 |
| chr20_-_8443425 | 0.87 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr4_+_6572364 | 0.81 |
ENSDART00000122574
|
ppp1r3aa
|
protein phosphatase 1, regulatory subunit 3Aa |
| chr24_+_41940299 | 0.81 |
ENSDART00000022349
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr17_+_45106706 | 0.80 |
ENSDART00000156971
|
GSTZ1
|
zgc:163014 |
| chr4_+_25630555 | 0.80 |
ENSDART00000133425
|
acot15
|
acyl-CoA thioesterase 15 |
| chr5_-_44843738 | 0.80 |
ENSDART00000003926
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr17_+_45106424 | 0.80 |
ENSDART00000157338
ENSDART00000155104 |
GSTZ1
|
zgc:163014 |
| chr19_-_30404096 | 0.80 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr10_+_38775959 | 0.79 |
ENSDART00000192990
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr21_-_41025340 | 0.78 |
ENSDART00000148231
|
plac8l1
|
PLAC8-like 1 |
| chr24_+_24086249 | 0.78 |
ENSDART00000002953
|
lipib
|
lipase, member Ib |
| chr20_-_48604621 | 0.78 |
ENSDART00000161769
ENSDART00000157871 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr7_+_17992455 | 0.75 |
ENSDART00000089211
ENSDART00000190881 |
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr5_-_71460556 | 0.74 |
ENSDART00000108804
|
brinp1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr20_-_33515890 | 0.73 |
ENSDART00000159987
|
si:dkey-65b13.13
|
si:dkey-65b13.13 |
| chr13_+_1430094 | 0.73 |
ENSDART00000169888
|
si:ch211-165e15.1
|
si:ch211-165e15.1 |
| chr24_+_24086491 | 0.73 |
ENSDART00000145092
|
lipib
|
lipase, member Ib |
| chr7_-_23971497 | 0.73 |
ENSDART00000173603
|
si:dkey-183c6.8
|
si:dkey-183c6.8 |
| chr3_+_17846890 | 0.67 |
ENSDART00000193384
|
znf385c
|
zinc finger protein 385C |
| chr21_+_9576176 | 0.66 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr24_-_28229618 | 0.65 |
ENSDART00000145290
|
bcl2a
|
BCL2, apoptosis regulator a |
| chr8_+_22859718 | 0.65 |
ENSDART00000134151
|
cacna1fb
|
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr16_-_13613475 | 0.65 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr5_-_5669879 | 0.65 |
ENSDART00000191963
|
CABZ01075628.1
|
|
| chr12_+_5209822 | 0.64 |
ENSDART00000152610
|
SLC35G1
|
si:ch211-197g18.2 |
| chr8_-_37263524 | 0.64 |
ENSDART00000061327
|
rh50
|
Rh50-like protein |
| chr9_-_30145080 | 0.62 |
ENSDART00000133746
|
abi3bpa
|
ABI family, member 3 (NESH) binding protein a |
| chr23_-_15878879 | 0.61 |
ENSDART00000010119
|
eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr5_-_65262260 | 0.60 |
ENSDART00000167886
|
col27a1b
|
collagen, type XXVII, alpha 1b |
| chr14_-_30490465 | 0.59 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr19_-_30403922 | 0.59 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr20_-_40367493 | 0.58 |
ENSDART00000075096
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr15_-_16679517 | 0.58 |
ENSDART00000177384
|
caln1
|
calneuron 1 |
| chr13_+_35746440 | 0.57 |
ENSDART00000187859
|
gpr75
|
G protein-coupled receptor 75 |
| chr14_-_30490763 | 0.57 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr21_-_39327223 | 0.57 |
ENSDART00000115097
|
aifm5
|
apoptosis-inducing factor, mitochondrion-associated, 5 |
| chr7_+_17096903 | 0.57 |
ENSDART00000145357
ENSDART00000132706 |
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr23_+_23183449 | 0.57 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr5_-_31928913 | 0.57 |
ENSDART00000142919
|
ssh1b
|
slingshot protein phosphatase 1b |
| chr16_+_51326237 | 0.56 |
ENSDART00000168525
|
CU469384.1
|
|
| chr18_-_41219790 | 0.56 |
ENSDART00000162387
ENSDART00000193753 |
zbtb38
|
zinc finger and BTB domain containing 38 |
| chr14_+_11457500 | 0.55 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr14_+_34068075 | 0.55 |
ENSDART00000135556
|
lonrf1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr21_-_40782393 | 0.53 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr25_-_16818978 | 0.53 |
ENSDART00000104140
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr6_+_43903209 | 0.51 |
ENSDART00000006435
|
gpr27
|
G protein-coupled receptor 27 |
| chr25_+_34581494 | 0.50 |
ENSDART00000167690
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr24_+_26039464 | 0.48 |
ENSDART00000131017
|
tnk2a
|
tyrosine kinase, non-receptor, 2a |
| chr15_-_16070731 | 0.48 |
ENSDART00000122099
|
dynll2a
|
dynein, light chain, LC8-type 2a |
| chr24_-_24281792 | 0.48 |
ENSDART00000146482
ENSDART00000018420 |
pdha1b
|
pyruvate dehydrogenase E1 alpha 1 subunit b |
| chr19_-_17658160 | 0.48 |
ENSDART00000151766
ENSDART00000170790 ENSDART00000186678 ENSDART00000188045 ENSDART00000176980 ENSDART00000166313 ENSDART00000188589 |
thrb
|
thyroid hormone receptor beta |
| chr20_-_35470891 | 0.47 |
ENSDART00000152993
ENSDART00000016090 |
pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr11_+_10548171 | 0.46 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr12_+_23850661 | 0.44 |
ENSDART00000152921
|
svila
|
supervillin a |
| chr7_+_62004048 | 0.44 |
ENSDART00000181818
|
smim20
|
small integral membrane protein 20 |
| chr11_+_26609110 | 0.44 |
ENSDART00000042322
|
map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr20_+_48100261 | 0.43 |
ENSDART00000158604
|
xkr5a
|
XK related 5a |
| chr17_-_22001303 | 0.43 |
ENSDART00000122190
|
slc22a7b.2
|
solute carrier family 22 (organic anion transporter), member 7b, tandem duplicate 2 |
| chr5_-_64823750 | 0.42 |
ENSDART00000140305
|
lix1
|
limb and CNS expressed 1 |
| chr10_-_17501528 | 0.41 |
ENSDART00000144847
|
slc2a11l
|
solute carrier family 2 (facilitated glucose transporter), member 11-like |
| chr1_-_46859398 | 0.40 |
ENSDART00000135795
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr15_+_25439106 | 0.40 |
ENSDART00000156252
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr21_-_2415808 | 0.40 |
ENSDART00000171179
|
si:ch211-241b2.5
|
si:ch211-241b2.5 |
| chr20_-_1314355 | 0.40 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr18_-_42333428 | 0.40 |
ENSDART00000034225
|
cntn5
|
contactin 5 |
| chr7_-_2116512 | 0.38 |
ENSDART00000098148
|
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr22_-_18116635 | 0.38 |
ENSDART00000005724
|
ncanb
|
neurocan b |
| chr7_-_15413188 | 0.37 |
ENSDART00000185980
|
si:ch211-106j24.1
|
si:ch211-106j24.1 |
| chr22_-_10354381 | 0.37 |
ENSDART00000092050
|
stab1
|
stabilin 1 |
| chr7_+_50464500 | 0.37 |
ENSDART00000191356
|
serinc4
|
serine incorporator 4 |
| chr5_+_21891305 | 0.37 |
ENSDART00000136788
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr20_-_1314537 | 0.36 |
ENSDART00000144288
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr1_+_52398205 | 0.32 |
ENSDART00000143225
|
si:ch211-217k17.9
|
si:ch211-217k17.9 |
| chr8_+_13389115 | 0.32 |
ENSDART00000184428
ENSDART00000154266 ENSDART00000049469 |
jak3
|
Janus kinase 3 (a protein tyrosine kinase, leukocyte) |
| chr3_+_49097775 | 0.32 |
ENSDART00000169185
|
zgc:123284
|
zgc:123284 |
| chr23_+_3731375 | 0.32 |
ENSDART00000141782
|
smim29
|
small integral membrane protein 29 |
| chr21_-_11820379 | 0.32 |
ENSDART00000126640
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr21_+_45819662 | 0.31 |
ENSDART00000193362
ENSDART00000184255 |
pitx1
|
paired-like homeodomain 1 |
| chr10_+_42462577 | 0.31 |
ENSDART00000133463
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr12_+_48216662 | 0.29 |
ENSDART00000187369
|
lrrc20
|
leucine rich repeat containing 20 |
| chr10_-_4375190 | 0.29 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr4_+_3438510 | 0.29 |
ENSDART00000155320
|
atxn7l1
|
ataxin 7-like 1 |
| chr7_-_32833153 | 0.28 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr25_+_32474031 | 0.27 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr21_-_24865217 | 0.27 |
ENSDART00000101136
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr19_+_43753995 | 0.27 |
ENSDART00000058504
|
SLC17A3
|
si:ch1073-513e17.1 |
| chr15_+_40792667 | 0.27 |
ENSDART00000193186
ENSDART00000184559 |
fat3a
|
FAT atypical cadherin 3a |
| chr1_+_51312752 | 0.27 |
ENSDART00000063938
|
mast1a
|
microtubule associated serine/threonine kinase 1a |
| chr9_+_23224761 | 0.26 |
ENSDART00000142008
|
map3k19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr13_-_22907260 | 0.26 |
ENSDART00000143097
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr3_-_31833266 | 0.24 |
ENSDART00000084932
ENSDART00000183617 |
map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr24_-_24146875 | 0.23 |
ENSDART00000173052
|
map7d2b
|
MAP7 domain containing 2b |
| chr19_+_43119698 | 0.22 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr2_-_31800521 | 0.22 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr5_-_50640171 | 0.22 |
ENSDART00000183353
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr18_+_33009828 | 0.22 |
ENSDART00000160100
|
olfcj1
|
olfactory receptor C family, j1 |
| chr11_-_25775447 | 0.21 |
ENSDART00000161301
|
plch2b
|
phospholipase C, eta 2b |
| chr21_+_25092300 | 0.21 |
ENSDART00000168505
|
dixdc1b
|
DIX domain containing 1b |
| chr8_+_30456161 | 0.20 |
ENSDART00000085894
|
pgm5
|
phosphoglucomutase 5 |
| chr5_+_51909740 | 0.18 |
ENSDART00000162541
|
thbs4a
|
thrombospondin 4a |
| chr14_-_32824380 | 0.18 |
ENSDART00000172791
ENSDART00000105745 |
inppl1b
|
inositol polyphosphate phosphatase-like 1b |
| chr19_+_30990129 | 0.18 |
ENSDART00000052169
ENSDART00000193376 |
sync
|
syncoilin, intermediate filament protein |
| chr18_+_37015185 | 0.18 |
ENSDART00000191305
|
sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr6_+_27315173 | 0.17 |
ENSDART00000108831
|
espnla
|
espin like a |
| chr6_+_515181 | 0.16 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr18_-_8030073 | 0.16 |
ENSDART00000151490
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr3_-_55139127 | 0.16 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr9_+_22466218 | 0.15 |
ENSDART00000131214
|
dgkg
|
diacylglycerol kinase, gamma |
| chr9_+_19304697 | 0.15 |
ENSDART00000040449
|
htr1fa
|
5-hydroxytryptamine (serotonin) receptor 1Fa |
| chr8_+_42629748 | 0.15 |
ENSDART00000075553
ENSDART00000135525 |
crybb2
|
crystallin, beta B2 |
| chr21_-_41369539 | 0.15 |
ENSDART00000187546
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr18_-_16315822 | 0.14 |
ENSDART00000136479
|
rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr14_-_30960470 | 0.13 |
ENSDART00000129989
|
si:ch211-191o15.6
|
si:ch211-191o15.6 |
| chr1_-_53750522 | 0.13 |
ENSDART00000190755
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr3_-_32831971 | 0.12 |
ENSDART00000075270
|
zgc:153733
|
zgc:153733 |
| chr7_+_57109214 | 0.12 |
ENSDART00000135068
ENSDART00000098412 |
enosf1
|
enolase superfamily member 1 |
| chr21_+_37436907 | 0.12 |
ENSDART00000182611
ENSDART00000076328 |
pgrmc1
|
progesterone receptor membrane component 1 |
| chr12_-_13729263 | 0.11 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr13_-_17723417 | 0.11 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr12_-_1951233 | 0.11 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr14_-_32884138 | 0.11 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr17_-_23673864 | 0.10 |
ENSDART00000104738
ENSDART00000128958 |
ptena
|
phosphatase and tensin homolog A |
| chr3_-_8765165 | 0.10 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr17_-_23674065 | 0.10 |
ENSDART00000104735
ENSDART00000182952 |
ptena
|
phosphatase and tensin homolog A |
| chr4_+_51551318 | 0.09 |
ENSDART00000190283
|
si:dkey-165e24.1
|
si:dkey-165e24.1 |
| chr21_-_5393125 | 0.09 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr3_+_43374571 | 0.09 |
ENSDART00000182497
|
zfand2a
|
zinc finger, AN1-type domain 2A |
| chr3_+_32118670 | 0.09 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr14_+_16765992 | 0.07 |
ENSDART00000140061
|
sqstm1
|
sequestosome 1 |
| chr17_+_15674052 | 0.06 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfe2l2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0015824 | proline transport(GO:0015824) |
| 0.4 | 1.2 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.3 | 1.0 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.3 | 1.0 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.3 | 1.4 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.3 | 0.8 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.2 | 1.5 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.2 | 2.3 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.2 | 1.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.2 | 1.0 | GO:0046874 | quinolinate biosynthetic process(GO:0019805) quinolinate metabolic process(GO:0046874) |
| 0.2 | 0.5 | GO:0043703 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 1.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 1.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.7 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.1 | 1.8 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 1.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 2.0 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 0.9 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 1.0 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.1 | 1.2 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.3 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.1 | 0.4 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.3 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.1 | 1.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.5 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 0.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 1.4 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 1.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 2.0 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.0 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 1.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 2.5 | GO:1903038 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.7 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.8 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.2 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 0.1 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.4 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.0 | 3.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 1.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.9 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 1.5 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 1.7 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.9 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.6 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 1.3 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 2.4 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.9 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 3.4 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.9 | GO:0007596 | blood coagulation(GO:0007596) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 3.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 2.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 1.0 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 1.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 3.0 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.5 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 1.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.0 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.3 | 0.8 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 1.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 2.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 0.5 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 1.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.8 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 3.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 2.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.5 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 1.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 2.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.3 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.1 | 3.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.9 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 2.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.7 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 2.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.8 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.2 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 1.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 1.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 2.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 2.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.9 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 2.5 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.8 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 2.4 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 0.9 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 2.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |