Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
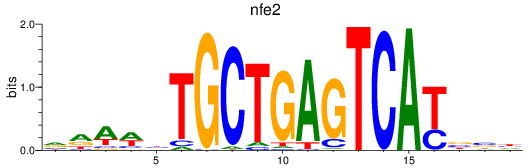
Results for nfe2
Z-value: 0.94
Transcription factors associated with nfe2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfe2
|
ENSDARG00000020606 | nuclear factor, erythroid 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfe2 | dr11_v1_chr23_-_36313431_36313431 | -0.10 | 6.8e-01 | Click! |
Activity profile of nfe2 motif
Sorted Z-values of nfe2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_30615901 | 3.20 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr22_-_16416882 | 2.35 |
ENSDART00000062749
|
cts12
|
cathepsin 12 |
| chr20_-_48604199 | 1.82 |
ENSDART00000161762
ENSDART00000170894 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr15_+_37559570 | 1.72 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr11_+_43401592 | 1.65 |
ENSDART00000112468
|
vipb
|
vasoactive intestinal peptide b |
| chr4_+_25630555 | 1.62 |
ENSDART00000133425
|
acot15
|
acyl-CoA thioesterase 15 |
| chr11_+_43401201 | 1.46 |
ENSDART00000190128
|
vipb
|
vasoactive intestinal peptide b |
| chr15_-_21877726 | 1.41 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr5_-_67471375 | 1.38 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr20_-_48604621 | 1.38 |
ENSDART00000161769
ENSDART00000157871 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr19_+_8506178 | 1.32 |
ENSDART00000189689
|
s100a10a
|
S100 calcium binding protein A10a |
| chr5_+_44805028 | 1.09 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr14_+_30279391 | 1.06 |
ENSDART00000172794
|
fgl1
|
fibrinogen-like 1 |
| chr3_-_55139127 | 1.05 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr2_+_9822319 | 1.01 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr23_-_29376859 | 0.99 |
ENSDART00000146411
|
sst6
|
somatostatin 6 |
| chr25_-_7999756 | 0.97 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr19_-_6873107 | 0.95 |
ENSDART00000124440
|
CABZ01029822.1
|
|
| chr18_+_2222447 | 0.93 |
ENSDART00000185927
|
pigbos1
|
PIGB opposite strand 1 |
| chr5_+_44805269 | 0.91 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr20_-_39271844 | 0.90 |
ENSDART00000192708
|
clu
|
clusterin |
| chr3_-_26017592 | 0.87 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr10_-_7974155 | 0.86 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr3_-_26017831 | 0.85 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr5_+_44804791 | 0.80 |
ENSDART00000122288
|
ctsla
|
cathepsin La |
| chr14_+_3507326 | 0.78 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr10_+_26747755 | 0.77 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr6_+_21395051 | 0.75 |
ENSDART00000017774
|
cacng5a
|
calcium channel, voltage-dependent, gamma subunit 5a |
| chr14_-_25985698 | 0.73 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr5_+_42064144 | 0.70 |
ENSDART00000035235
|
si:ch211-202a12.4
|
si:ch211-202a12.4 |
| chr9_-_23253870 | 0.70 |
ENSDART00000143657
ENSDART00000169911 |
acmsd
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr12_+_20587179 | 0.69 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr20_-_16906623 | 0.68 |
ENSDART00000012859
ENSDART00000171628 |
psma6b
|
proteasome subunit alpha 6b |
| chr7_+_57109214 | 0.66 |
ENSDART00000135068
ENSDART00000098412 |
enosf1
|
enolase superfamily member 1 |
| chr16_+_13883872 | 0.61 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr9_-_33328948 | 0.60 |
ENSDART00000006948
|
rpl8
|
ribosomal protein L8 |
| chr25_-_8625601 | 0.59 |
ENSDART00000155280
|
GDPGP1
|
zgc:153343 |
| chr16_-_41804606 | 0.57 |
ENSDART00000134718
|
psmc4
|
proteasome 26S subunit, ATPase 4 |
| chr14_+_34068075 | 0.56 |
ENSDART00000135556
|
lonrf1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr15_-_47929455 | 0.56 |
ENSDART00000064462
|
psma6l
|
proteasome subunit alpha 6, like |
| chr23_+_9560797 | 0.54 |
ENSDART00000180014
|
adrm1
|
adhesion regulating molecule 1 |
| chr20_-_39103119 | 0.54 |
ENSDART00000143379
|
rcan2
|
regulator of calcineurin 2 |
| chr21_-_5393125 | 0.53 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr25_+_32474031 | 0.53 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr2_-_6115688 | 0.52 |
ENSDART00000081663
|
prdx1
|
peroxiredoxin 1 |
| chr12_+_48216662 | 0.52 |
ENSDART00000187369
|
lrrc20
|
leucine rich repeat containing 20 |
| chr19_-_32600823 | 0.51 |
ENSDART00000134149
ENSDART00000187858 |
zgc:91944
|
zgc:91944 |
| chr22_-_13857729 | 0.51 |
ENSDART00000177971
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr8_-_18010735 | 0.49 |
ENSDART00000125014
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr25_-_35182347 | 0.48 |
ENSDART00000115210
|
ano9a
|
anoctamin 9a |
| chr19_-_6385594 | 0.46 |
ENSDART00000104950
|
atp1a3a
|
ATPase Na+/K+ transporting subunit alpha 3a |
| chr13_+_25396896 | 0.44 |
ENSDART00000041257
|
gsto2
|
glutathione S-transferase omega 2 |
| chr10_+_29265463 | 0.44 |
ENSDART00000155390
|
crebzf
|
CREB/ATF bZIP transcription factor |
| chr20_-_19422496 | 0.43 |
ENSDART00000143658
|
si:ch211-278j3.3
|
si:ch211-278j3.3 |
| chr10_+_31222433 | 0.43 |
ENSDART00000185080
|
tmem218
|
transmembrane protein 218 |
| chr18_+_1703984 | 0.43 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr7_+_17096903 | 0.42 |
ENSDART00000145357
ENSDART00000132706 |
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr23_+_44546319 | 0.42 |
ENSDART00000139088
ENSDART00000056935 |
trappc1
|
trafficking protein particle complex 1 |
| chr16_+_46410520 | 0.42 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr5_-_44843738 | 0.41 |
ENSDART00000003926
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr14_-_17588345 | 0.40 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr1_+_54655160 | 0.39 |
ENSDART00000190319
|
si:ch211-202h22.7
|
si:ch211-202h22.7 |
| chr4_-_8902406 | 0.39 |
ENSDART00000192962
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr13_+_7241170 | 0.38 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr13_+_25397098 | 0.38 |
ENSDART00000132953
|
gsto2
|
glutathione S-transferase omega 2 |
| chr4_-_20292821 | 0.37 |
ENSDART00000136069
ENSDART00000192504 |
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr25_-_11378623 | 0.37 |
ENSDART00000166586
|
enc2
|
ectodermal-neural cortex 2 |
| chr4_+_1757462 | 0.37 |
ENSDART00000032460
|
med21
|
mediator complex subunit 21 |
| chr4_+_5317483 | 0.36 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr21_-_25756119 | 0.35 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr23_-_44546124 | 0.35 |
ENSDART00000126779
ENSDART00000024082 |
psmb6
|
proteasome subunit beta 6 |
| chr19_-_41404870 | 0.33 |
ENSDART00000167772
|
sem1
|
SEM1, 26S proteasome complex subunit |
| chr9_+_6937569 | 0.33 |
ENSDART00000146383
|
mfsd9
|
major facilitator superfamily domain containing 9 |
| chr19_+_43119698 | 0.33 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr12_-_18898413 | 0.33 |
ENSDART00000181281
ENSDART00000121866 |
desi1b
|
desumoylating isopeptidase 1b |
| chr8_+_4434422 | 0.33 |
ENSDART00000145184
|
srrd
|
SRR1 domain containing |
| chr17_-_11439815 | 0.33 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr2_-_6115389 | 0.33 |
ENSDART00000134921
|
prdx1
|
peroxiredoxin 1 |
| chr17_-_7861219 | 0.32 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr11_+_36777613 | 0.32 |
ENSDART00000065644
ENSDART00000158151 |
parapinopsinb
|
parapinopsin b |
| chr5_-_5669879 | 0.32 |
ENSDART00000191963
|
CABZ01075628.1
|
|
| chr15_-_28677725 | 0.32 |
ENSDART00000060255
|
blmh
|
bleomycin hydrolase |
| chr17_-_12966907 | 0.31 |
ENSDART00000022874
|
psma6a
|
proteasome subunit alpha 6a |
| chr18_-_2222128 | 0.31 |
ENSDART00000171402
|
pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr23_+_9560991 | 0.31 |
ENSDART00000081433
ENSDART00000131594 ENSDART00000130069 ENSDART00000138601 |
adrm1
|
adhesion regulating molecule 1 |
| chr7_-_19043401 | 0.31 |
ENSDART00000158796
|
fcer1g
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
| chr9_+_11202552 | 0.30 |
ENSDART00000151931
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr21_-_11286483 | 0.29 |
ENSDART00000074845
|
rtkn2b
|
rhotekin 2b |
| chr15_+_25438714 | 0.28 |
ENSDART00000154164
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr15_-_1590858 | 0.28 |
ENSDART00000081875
|
nnr
|
nanor |
| chr25_-_35045250 | 0.27 |
ENSDART00000156508
ENSDART00000126590 |
zgc:114046
|
zgc:114046 |
| chr25_+_29474583 | 0.27 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr25_+_3549401 | 0.26 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr1_+_6640437 | 0.26 |
ENSDART00000147638
|
si:ch211-93g23.2
|
si:ch211-93g23.2 |
| chr17_-_23673864 | 0.25 |
ENSDART00000104738
ENSDART00000128958 |
ptena
|
phosphatase and tensin homolog A |
| chr11_-_29936755 | 0.24 |
ENSDART00000145251
|
pir
|
pirin |
| chr12_+_1455147 | 0.24 |
ENSDART00000018752
|
cops3
|
COP9 signalosome subunit 3 |
| chr20_-_35557902 | 0.24 |
ENSDART00000134753
|
adgrf7
|
adhesion G protein-coupled receptor F7 |
| chr22_+_19528851 | 0.23 |
ENSDART00000145079
|
si:dkey-78l4.13
|
si:dkey-78l4.13 |
| chr3_+_58092212 | 0.22 |
ENSDART00000156059
|
si:ch211-256e16.7
|
si:ch211-256e16.7 |
| chr2_-_9544161 | 0.22 |
ENSDART00000124425
|
slc25a24l
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24, like |
| chr17_-_23674065 | 0.22 |
ENSDART00000104735
ENSDART00000182952 |
ptena
|
phosphatase and tensin homolog A |
| chr14_-_33312407 | 0.21 |
ENSDART00000135700
|
sept6
|
septin 6 |
| chr6_+_53288018 | 0.21 |
ENSDART00000170319
|
cysltr3
|
cysteinyl leukotriene receptor 3 |
| chr24_+_24086249 | 0.21 |
ENSDART00000002953
|
lipib
|
lipase, member Ib |
| chr23_-_15878879 | 0.21 |
ENSDART00000010119
|
eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr17_+_7524389 | 0.20 |
ENSDART00000067579
|
klhl10b.2
|
kelch-like family member 10b, tandem duplicate 2 |
| chr24_-_9689915 | 0.20 |
ENSDART00000185972
ENSDART00000093046 |
uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr6_-_425378 | 0.19 |
ENSDART00000192190
|
fam83fb
|
family with sequence similarity 83, member Fb |
| chr8_+_18010568 | 0.19 |
ENSDART00000121984
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr8_+_18010978 | 0.19 |
ENSDART00000039887
ENSDART00000144532 |
ssbp3b
|
single stranded DNA binding protein 3b |
| chr3_-_34816893 | 0.19 |
ENSDART00000084448
ENSDART00000154696 |
psmd11a
|
proteasome 26S subunit, non-ATPase 11a |
| chr15_+_25439106 | 0.19 |
ENSDART00000156252
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr7_+_57108823 | 0.18 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr16_+_9762261 | 0.18 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr17_+_33418475 | 0.18 |
ENSDART00000169145
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr21_-_45349548 | 0.17 |
ENSDART00000008454
|
skp1
|
S-phase kinase-associated protein 1 |
| chr20_-_19532531 | 0.17 |
ENSDART00000125830
ENSDART00000179566 ENSDART00000184193 |
eif2b4
BX276101.2
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr18_+_33009828 | 0.17 |
ENSDART00000160100
|
olfcj1
|
olfactory receptor C family, j1 |
| chr25_+_29474982 | 0.17 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr8_+_24747865 | 0.17 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr15_-_26552652 | 0.15 |
ENSDART00000152336
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr7_+_1045637 | 0.15 |
ENSDART00000111531
|
epdl1
|
ependymin-like 1 |
| chr19_-_10478229 | 0.15 |
ENSDART00000142937
|
si:ch211-171h4.5
|
si:ch211-171h4.5 |
| chr14_-_38872536 | 0.14 |
ENSDART00000184315
ENSDART00000127479 |
gsr
|
glutathione reductase |
| chr18_-_28938912 | 0.14 |
ENSDART00000136201
|
si:ch211-174j14.2
|
si:ch211-174j14.2 |
| chr4_-_60780423 | 0.14 |
ENSDART00000162632
|
si:dkey-254e13.6
|
si:dkey-254e13.6 |
| chr1_-_52497834 | 0.14 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr24_+_24086491 | 0.14 |
ENSDART00000145092
|
lipib
|
lipase, member Ib |
| chr7_-_47282853 | 0.13 |
ENSDART00000169299
|
si:ch211-186j3.3
|
si:ch211-186j3.3 |
| chr25_+_10410620 | 0.13 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr1_+_51039558 | 0.13 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr6_+_9421279 | 0.12 |
ENSDART00000161036
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr22_+_9027884 | 0.12 |
ENSDART00000171839
|
si:ch211-213a13.2
|
si:ch211-213a13.2 |
| chr23_+_9508538 | 0.11 |
ENSDART00000010697
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr22_+_1110855 | 0.11 |
ENSDART00000168251
|
si:ch1073-181h11.1
|
si:ch1073-181h11.1 |
| chr17_-_22001303 | 0.11 |
ENSDART00000122190
|
slc22a7b.2
|
solute carrier family 22 (organic anion transporter), member 7b, tandem duplicate 2 |
| chr14_+_28464736 | 0.10 |
ENSDART00000019003
|
psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr14_-_7207961 | 0.10 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr3_-_21280373 | 0.09 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr5_+_42912966 | 0.09 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr8_-_21056012 | 0.09 |
ENSDART00000142852
|
zgc:112962
|
zgc:112962 |
| chr19_-_10432134 | 0.09 |
ENSDART00000081440
|
il11b
|
interleukin 11b |
| chr9_-_34509997 | 0.09 |
ENSDART00000169114
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr1_-_52498146 | 0.08 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr3_+_17951790 | 0.07 |
ENSDART00000164663
|
aclya
|
ATP citrate lyase a |
| chr3_+_30257582 | 0.07 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr16_-_27640995 | 0.06 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr1_+_48256950 | 0.06 |
ENSDART00000171182
|
SORCS3
|
sortilin related VPS10 domain containing receptor 3 |
| chr8_+_20825987 | 0.06 |
ENSDART00000133309
|
si:ch211-133l5.4
|
si:ch211-133l5.4 |
| chr21_+_8341774 | 0.06 |
ENSDART00000129749
ENSDART00000055325 ENSDART00000133804 |
psmb7
|
proteasome subunit beta 7 |
| chr24_-_24281792 | 0.06 |
ENSDART00000146482
ENSDART00000018420 |
pdha1b
|
pyruvate dehydrogenase E1 alpha 1 subunit b |
| chr16_+_24721914 | 0.04 |
ENSDART00000109459
|
smg9
|
smg9 nonsense mediated mRNA decay factor |
| chr4_-_36476889 | 0.04 |
ENSDART00000163956
|
si:ch211-263l8.1
|
si:ch211-263l8.1 |
| chr11_+_13630107 | 0.04 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr4_-_50930346 | 0.04 |
ENSDART00000184245
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr14_-_38873095 | 0.03 |
ENSDART00000047050
ENSDART00000173285 ENSDART00000147521 |
gsr
|
glutathione reductase |
| chr9_+_26199674 | 0.03 |
ENSDART00000139510
|
si:dkey-111i23.1
|
si:dkey-111i23.1 |
| chr13_-_51973562 | 0.02 |
ENSDART00000166701
|
slc30a10
|
solute carrier family 30, member 10 |
| chr9_+_22466218 | 0.01 |
ENSDART00000131214
|
dgkg
|
diacylglycerol kinase, gamma |
| chr4_+_76442674 | 0.01 |
ENSDART00000164825
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr3_+_52684556 | 0.01 |
ENSDART00000139037
|
pgls
|
6-phosphogluconolactonase |
| chr13_+_42309688 | 0.00 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfe2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0070459 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.6 | 1.7 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.3 | 1.4 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.2 | 0.5 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.1 | 2.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.4 | GO:0046351 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.7 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) quinolinate metabolic process(GO:0046874) |
| 0.1 | 0.6 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.5 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.1 | 0.7 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.6 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.8 | GO:0071450 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.3 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.1 | 2.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 1.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.5 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.3 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.8 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.3 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.2 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.5 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 2.2 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.8 | GO:0050868 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.4 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.7 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 0.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.3 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 0.9 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 1.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 3.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.6 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 5.1 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 2.9 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.3 | 1.4 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.3 | 0.8 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.2 | 0.5 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.1 | 0.7 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 3.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 3.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.4 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.4 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.5 | GO:0051800 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 0.3 | GO:0019767 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.1 | 0.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.5 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 1.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.4 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.6 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 4.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 1.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |