Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
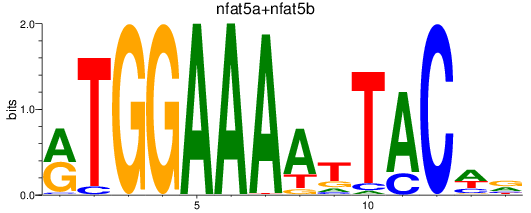
Results for nfat5a+nfat5b
Z-value: 0.55
Transcription factors associated with nfat5a+nfat5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfat5a
|
ENSDARG00000020872 | nuclear factor of activated T cells 5a |
|
nfat5b
|
ENSDARG00000102556 | nuclear factor of activated T cells 5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfat5b | dr11_v1_chr7_+_67325933_67325933 | 0.54 | 1.8e-02 | Click! |
| nfat5a | dr11_v1_chr25_-_37121335_37121335 | 0.25 | 3.0e-01 | Click! |
Activity profile of nfat5a+nfat5b motif
Sorted Z-values of nfat5a+nfat5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_612594 | 1.11 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr23_-_27875140 | 1.01 |
ENSDART00000143662
|
ankrd33aa
|
ankyrin repeat domain 33Aa |
| chr21_+_6291027 | 0.88 |
ENSDART00000180467
ENSDART00000184952 ENSDART00000184006 |
fnbp1b
|
formin binding protein 1b |
| chr8_+_1843135 | 0.87 |
ENSDART00000141452
|
snap29
|
synaptosomal-associated protein 29 |
| chr2_-_37960688 | 0.86 |
ENSDART00000055565
|
cbln14
|
cerebellin 14 |
| chr16_+_45746549 | 0.85 |
ENSDART00000190403
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr20_-_53624645 | 0.83 |
ENSDART00000083427
ENSDART00000152920 |
slc25a29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr2_-_26499842 | 0.82 |
ENSDART00000186929
|
BX569796.1
|
|
| chr2_-_5502256 | 0.82 |
ENSDART00000193037
|
saga
|
S-antigen; retina and pineal gland (arrestin) a |
| chr10_+_22782522 | 0.80 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr3_-_5067585 | 0.76 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr18_+_45792035 | 0.75 |
ENSDART00000135045
|
abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr25_-_35136267 | 0.75 |
ENSDART00000109751
|
zgc:173585
|
zgc:173585 |
| chr18_-_898870 | 0.75 |
ENSDART00000151777
ENSDART00000062654 |
parp6a
|
poly (ADP-ribose) polymerase family, member 6a |
| chr1_+_41666611 | 0.75 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr5_+_13521081 | 0.74 |
ENSDART00000171975
|
si:ch211-230g14.6
|
si:ch211-230g14.6 |
| chr2_-_37888429 | 0.73 |
ENSDART00000183277
|
mbl2
|
mannose binding lectin 2 |
| chr1_+_54194801 | 0.67 |
ENSDART00000186802
|
CABZ01067150.1
|
|
| chr7_+_39444843 | 0.66 |
ENSDART00000143999
ENSDART00000173554 ENSDART00000173698 ENSDART00000173754 ENSDART00000144075 ENSDART00000138192 ENSDART00000145457 ENSDART00000141750 ENSDART00000103056 ENSDART00000142946 ENSDART00000173748 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr5_-_38384289 | 0.66 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr13_+_33024547 | 0.65 |
ENSDART00000057377
|
arg2
|
arginase 2 |
| chr17_-_33414781 | 0.65 |
ENSDART00000142203
ENSDART00000034638 |
ccdc28a
|
coiled-coil domain containing 28A |
| chr17_+_36627099 | 0.64 |
ENSDART00000154104
|
impg1b
|
interphotoreceptor matrix proteoglycan 1b |
| chr9_+_7909490 | 0.59 |
ENSDART00000133063
ENSDART00000109288 |
myo16
|
myosin XVI |
| chr21_+_5494561 | 0.56 |
ENSDART00000169728
|
ly6m3
|
lymphocyte antigen 6 family member M3 |
| chr9_-_11263228 | 0.55 |
ENSDART00000113847
|
chpfa
|
chondroitin polymerizing factor a |
| chr5_-_49012569 | 0.55 |
ENSDART00000184690
|
si:dkey-172m14.2
|
si:dkey-172m14.2 |
| chr17_-_277046 | 0.54 |
ENSDART00000182587
|
LO018437.1
|
|
| chr2_+_33541928 | 0.49 |
ENSDART00000162852
|
BX548164.1
|
|
| chr3_+_22059066 | 0.48 |
ENSDART00000155739
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr17_-_43012390 | 0.46 |
ENSDART00000155615
|
atg2b
|
autophagy related 2B |
| chr18_+_9641210 | 0.43 |
ENSDART00000182024
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr5_-_63109232 | 0.42 |
ENSDART00000115128
|
usp2b
|
ubiquitin specific peptidase 2b |
| chr17_+_33415319 | 0.40 |
ENSDART00000140805
ENSDART00000025501 ENSDART00000146447 |
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr2_+_55982300 | 0.39 |
ENSDART00000183903
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr7_+_39624728 | 0.39 |
ENSDART00000173847
ENSDART00000173845 |
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr20_-_1174266 | 0.38 |
ENSDART00000023304
|
gabrr2b
|
gamma-aminobutyric acid (GABA) A receptor, rho 2b |
| chr5_-_49011912 | 0.38 |
ENSDART00000097450
|
si:dkey-172m14.2
|
si:dkey-172m14.2 |
| chr8_+_7144066 | 0.37 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr23_-_26522760 | 0.37 |
ENSDART00000142417
ENSDART00000135606 ENSDART00000122668 |
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr16_+_2469889 | 0.36 |
ENSDART00000126759
|
cnih4
|
cornichon family AMPA receptor auxiliary protein 4 |
| chr2_-_24369087 | 0.36 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr12_-_30549022 | 0.35 |
ENSDART00000102474
|
zgc:158404
|
zgc:158404 |
| chr24_-_20656773 | 0.35 |
ENSDART00000190883
ENSDART00000193896 |
nktr
|
natural killer cell triggering receptor |
| chr3_+_40856095 | 0.34 |
ENSDART00000143207
|
mmd2a
|
monocyte to macrophage differentiation-associated 2a |
| chr21_+_22840246 | 0.34 |
ENSDART00000151621
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr6_+_40992883 | 0.33 |
ENSDART00000076061
|
tgfa
|
transforming growth factor, alpha |
| chr10_+_4807913 | 0.33 |
ENSDART00000186099
|
palm2
|
paralemmin 2 |
| chr24_-_28229618 | 0.33 |
ENSDART00000145290
|
bcl2a
|
BCL2, apoptosis regulator a |
| chr11_-_30611814 | 0.33 |
ENSDART00000089803
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr23_+_26102028 | 0.32 |
ENSDART00000139488
|
si:dkey-78k11.4
|
si:dkey-78k11.4 |
| chr13_-_11667661 | 0.32 |
ENSDART00000102411
|
dctn1b
|
dynactin 1b |
| chr1_+_2712956 | 0.31 |
ENSDART00000126093
|
gpc6a
|
glypican 6a |
| chr5_-_62940851 | 0.31 |
ENSDART00000137052
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr17_+_33311784 | 0.30 |
ENSDART00000156229
|
si:ch211-132f19.7
|
si:ch211-132f19.7 |
| chr4_+_57093908 | 0.30 |
ENSDART00000170198
|
si:ch211-238e22.5
|
si:ch211-238e22.5 |
| chr5_-_13353810 | 0.30 |
ENSDART00000146136
ENSDART00000099602 ENSDART00000004539 |
ube2l3b
|
ubiquitin-conjugating enzyme E2L 3b |
| chr3_-_30434016 | 0.30 |
ENSDART00000150958
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr12_-_17147473 | 0.29 |
ENSDART00000106035
|
stambpl1
|
STAM binding protein-like 1 |
| chr2_-_42492201 | 0.29 |
ENSDART00000180762
ENSDART00000009093 |
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr21_+_2742823 | 0.29 |
ENSDART00000189004
|
CABZ01073424.2
|
|
| chr15_-_18209672 | 0.28 |
ENSDART00000141508
ENSDART00000136280 |
btr16
|
bloodthirsty-related gene family, member 16 |
| chr7_-_2285314 | 0.28 |
ENSDART00000153614
|
si:dkey-187j14.6
|
si:dkey-187j14.6 |
| chr4_+_35594200 | 0.25 |
ENSDART00000193902
|
si:dkeyp-4c4.2
|
si:dkeyp-4c4.2 |
| chr19_+_32807236 | 0.25 |
ENSDART00000008698
|
stk3
|
serine/threonine kinase 3 (STE20 homolog, yeast) |
| chr19_-_8748571 | 0.24 |
ENSDART00000031173
|
rps27.1
|
ribosomal protein S27, isoform 1 |
| chr9_+_310331 | 0.23 |
ENSDART00000172446
ENSDART00000187731 ENSDART00000193970 |
stac3
|
SH3 and cysteine rich domain 3 |
| chr19_-_36234185 | 0.23 |
ENSDART00000186003
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr10_+_2981090 | 0.23 |
ENSDART00000111830
|
zfyve16
|
zinc finger, FYVE domain containing 16 |
| chr7_-_29115772 | 0.22 |
ENSDART00000076386
|
fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr4_+_76722754 | 0.21 |
ENSDART00000153867
|
ms4a17a.3
|
membrane-spanning 4-domains, subfamily A, member 17A.3 |
| chr1_+_19616799 | 0.21 |
ENSDART00000143077
|
fbxo10
|
F-box protein 10 |
| chr13_+_22104298 | 0.21 |
ENSDART00000115393
|
si:dkey-174i8.1
|
si:dkey-174i8.1 |
| chr5_-_40040488 | 0.21 |
ENSDART00000157755
|
cds1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr2_-_36925561 | 0.21 |
ENSDART00000187690
|
map1sb
|
microtubule-associated protein 1Sb |
| chr7_-_2285499 | 0.20 |
ENSDART00000182211
|
si:dkey-187j14.6
|
si:dkey-187j14.6 |
| chr2_-_34483597 | 0.20 |
ENSDART00000133224
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr2_-_24348948 | 0.19 |
ENSDART00000136559
|
ano8a
|
anoctamin 8a |
| chr9_+_53725128 | 0.19 |
ENSDART00000169062
|
cnmd
|
chondromodulin |
| chr20_-_40720458 | 0.19 |
ENSDART00000153151
ENSDART00000061261 ENSDART00000138569 |
cx43
|
connexin 43 |
| chr9_+_53725294 | 0.19 |
ENSDART00000165991
|
cnmd
|
chondromodulin |
| chr10_+_44955106 | 0.18 |
ENSDART00000185837
|
il1b
|
interleukin 1, beta |
| chr16_+_33902006 | 0.18 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr23_-_45897900 | 0.18 |
ENSDART00000149263
|
ftr92
|
finTRIM family, member 92 |
| chr5_-_42904329 | 0.17 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr20_-_29787192 | 0.17 |
ENSDART00000125348
ENSDART00000048759 |
id2b
|
inhibitor of DNA binding 2b |
| chr10_+_15255012 | 0.16 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr1_+_56502706 | 0.16 |
ENSDART00000188665
|
CABZ01059403.1
|
|
| chr10_+_21722892 | 0.16 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr2_-_24348642 | 0.15 |
ENSDART00000181739
|
ano8a
|
anoctamin 8a |
| chr17_+_15559046 | 0.14 |
ENSDART00000187126
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr10_-_40968095 | 0.14 |
ENSDART00000184104
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr21_+_44557006 | 0.14 |
ENSDART00000170159
|
cmc4
|
C-x(9)-C motif containing 4 homolog (S. cerevisiae) |
| chr5_-_9948497 | 0.14 |
ENSDART00000183196
|
tor4ab
|
torsin family 4, member Ab |
| chr6_+_6802142 | 0.14 |
ENSDART00000065566
|
dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr6_+_58607488 | 0.14 |
ENSDART00000182648
|
helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr6_-_19381993 | 0.13 |
ENSDART00000164114
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr7_+_29009161 | 0.13 |
ENSDART00000166458
|
CR376738.1
|
|
| chr24_-_20599781 | 0.13 |
ENSDART00000179664
ENSDART00000141823 |
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr10_-_32465462 | 0.13 |
ENSDART00000134056
|
uvrag
|
UV radiation resistance associated gene |
| chr6_+_612330 | 0.13 |
ENSDART00000166872
ENSDART00000191758 |
kynu
|
kynureninase |
| chr25_-_13842618 | 0.12 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr24_-_19719240 | 0.12 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr5_+_13353866 | 0.12 |
ENSDART00000099611
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr23_+_28770225 | 0.11 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr21_-_14762944 | 0.11 |
ENSDART00000114096
|
arrdc1b
|
arrestin domain containing 1b |
| chr12_-_33859950 | 0.11 |
ENSDART00000131181
|
sema4gb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Gb |
| chr4_-_52504961 | 0.11 |
ENSDART00000163123
|
si:dkey-78o7.3
|
si:dkey-78o7.3 |
| chr11_-_30612166 | 0.11 |
ENSDART00000177984
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr5_-_37900350 | 0.11 |
ENSDART00000084839
ENSDART00000084841 ENSDART00000133437 |
tmprss13b
|
transmembrane protease, serine 13b |
| chr10_-_42882215 | 0.10 |
ENSDART00000180580
ENSDART00000187374 |
FP236542.1
|
|
| chr5_-_19988772 | 0.10 |
ENSDART00000086881
|
slc6a4b
|
solute carrier family 6 (neurotransmitter transporter), member 4b |
| chr14_-_25599002 | 0.10 |
ENSDART00000040955
|
slc25a48
|
solute carrier family 25, member 48 |
| chr20_-_487783 | 0.09 |
ENSDART00000162218
|
col10a1b
|
collagen, type X, alpha 1b |
| chr10_-_37072776 | 0.09 |
ENSDART00000110835
|
myo18aa
|
myosin XVIIIAa |
| chr14_+_23970818 | 0.09 |
ENSDART00000123338
ENSDART00000124944 |
kif3a
|
kinesin family member 3A |
| chr2_-_41518340 | 0.09 |
ENSDART00000130830
|
otomp
|
otolith matrix protein |
| chr23_+_36771593 | 0.08 |
ENSDART00000078240
|
march9
|
membrane-associated ring finger (C3HC4) 9 |
| chr12_+_34891529 | 0.08 |
ENSDART00000015643
|
tbcc
|
tubulin folding cofactor C |
| chr1_+_54124209 | 0.08 |
ENSDART00000187730
|
LO017722.1
|
|
| chr3_-_2033105 | 0.07 |
ENSDART00000135249
|
si:dkey-88j15.3
|
si:dkey-88j15.3 |
| chr18_+_26895994 | 0.07 |
ENSDART00000098347
|
ch25hl1.2
|
cholesterol 25-hydroxylase like 1, tandem duplicate 2 |
| chr4_+_47751483 | 0.07 |
ENSDART00000160043
|
si:ch211-196f19.1
|
si:ch211-196f19.1 |
| chr24_-_21674950 | 0.07 |
ENSDART00000123216
ENSDART00000046211 |
lnx2a
|
ligand of numb-protein X 2a |
| chr20_-_23852174 | 0.06 |
ENSDART00000122414
|
si:dkey-15j16.6
|
si:dkey-15j16.6 |
| chr3_-_29688370 | 0.06 |
ENSDART00000151147
ENSDART00000151679 |
si:ch73-233k15.2
|
si:ch73-233k15.2 |
| chr4_-_53047883 | 0.06 |
ENSDART00000157496
|
si:dkey-201g16.5
|
si:dkey-201g16.5 |
| chr15_-_25099679 | 0.06 |
ENSDART00000154628
|
rflnb
|
refilin B |
| chr12_-_17024079 | 0.05 |
ENSDART00000129027
|
ifit11
|
interferon-induced protein with tetratricopeptide repeats 11 |
| chr7_+_48667081 | 0.05 |
ENSDART00000083473
|
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr2_+_3881000 | 0.05 |
ENSDART00000081897
|
mpp7b
|
membrane protein, palmitoylated 7b (MAGUK p55 subfamily member 7) |
| chr16_+_43077909 | 0.05 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
| chr22_-_32360464 | 0.05 |
ENSDART00000148886
|
pcbp4
|
poly(rC) binding protein 4 |
| chr2_+_55982940 | 0.05 |
ENSDART00000097753
ENSDART00000097751 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr9_+_21243985 | 0.05 |
ENSDART00000131586
|
si:rp71-68n21.12
|
si:rp71-68n21.12 |
| chr13_+_1015749 | 0.05 |
ENSDART00000190982
|
prokr1b
|
prokineticin receptor 1b |
| chr13_-_41155472 | 0.05 |
ENSDART00000160588
|
si:dkeyp-86d6.2
|
si:dkeyp-86d6.2 |
| chr19_-_4785734 | 0.05 |
ENSDART00000113088
|
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr3_+_50172452 | 0.04 |
ENSDART00000191224
|
epn3a
|
epsin 3a |
| chr20_-_52631998 | 0.04 |
ENSDART00000145283
ENSDART00000139072 |
si:ch211-221n20.1
|
si:ch211-221n20.1 |
| chr13_+_23623346 | 0.04 |
ENSDART00000057619
|
il22ra2
|
interleukin 22 receptor, alpha 2 |
| chr9_+_1352603 | 0.04 |
ENSDART00000146016
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr21_+_15435946 | 0.04 |
ENSDART00000137491
|
si:dkey-11o15.10
|
si:dkey-11o15.10 |
| chr16_+_29303971 | 0.04 |
ENSDART00000087149
|
hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr3_+_32080403 | 0.03 |
ENSDART00000124943
|
aldh16a1
|
aldehyde dehydrogenase 16 family, member A1 |
| chr19_-_25149034 | 0.03 |
ENSDART00000148432
ENSDART00000175266 |
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr18_+_41772474 | 0.03 |
ENSDART00000097547
|
trpc6b
|
transient receptor potential cation channel, subfamily C, member 6b |
| chr2_-_37862380 | 0.02 |
ENSDART00000186005
|
si:ch211-284o19.8
|
si:ch211-284o19.8 |
| chr22_+_1680628 | 0.02 |
ENSDART00000166672
|
si:dkey-1b17.6
|
si:dkey-1b17.6 |
| chr15_+_37950556 | 0.02 |
ENSDART00000153947
|
si:dkey-238d18.12
|
si:dkey-238d18.12 |
| chr19_+_43885770 | 0.02 |
ENSDART00000135599
|
lypla2
|
lysophospholipase II |
| chr1_-_493218 | 0.02 |
ENSDART00000031635
|
ercc5
|
excision repair cross-complementation group 5 |
| chr4_-_58883559 | 0.02 |
ENSDART00000164546
|
si:dkey-28i19.4
|
si:dkey-28i19.4 |
| chr1_-_29658721 | 0.01 |
ENSDART00000132063
|
si:dkey-1h24.6
|
si:dkey-1h24.6 |
| chr7_-_56793739 | 0.01 |
ENSDART00000082842
|
si:ch211-146m13.3
|
si:ch211-146m13.3 |
| chr10_+_21730585 | 0.01 |
ENSDART00000188576
|
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr14_-_33218418 | 0.00 |
ENSDART00000163046
|
si:dkey-31j3.11
|
si:dkey-31j3.11 |
| chr6_+_51932563 | 0.00 |
ENSDART00000181778
|
angpt4
|
angiopoietin 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfat5a+nfat5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.2 | 0.9 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.1 | 0.9 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.1 | 0.3 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 0.6 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.1 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.8 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.8 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.5 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.2 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.4 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.3 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.7 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 0.1 | GO:0006837 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.3 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.4 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.3 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.8 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.4 | GO:0072348 | sulfur compound transport(GO:0072348) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.4 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.0 | 0.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 0.9 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 0.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.5 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.7 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.2 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0015222 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |