Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
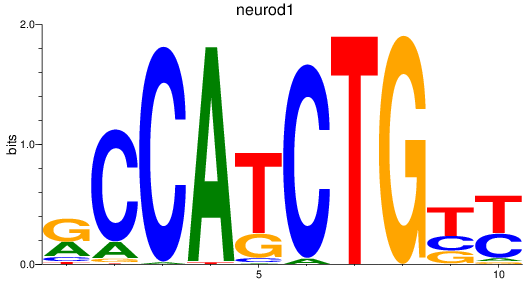
Results for neurod1
Z-value: 1.75
Transcription factors associated with neurod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
neurod1
|
ENSDARG00000019566 | neuronal differentiation 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| neurod1 | dr11_v1_chr9_-_44295071_44295071 | 0.59 | 8.1e-03 | Click! |
Activity profile of neurod1 motif
Sorted Z-values of neurod1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_51065048 | 14.72 |
ENSDART00000168085
|
CABZ01078663.1
|
|
| chr25_+_37130450 | 7.56 |
ENSDART00000183358
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr3_-_50443607 | 7.03 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr8_+_21376290 | 6.52 |
ENSDART00000136765
|
ela2
|
elastase 2 |
| chr12_+_46967789 | 6.48 |
ENSDART00000114866
|
oat
|
ornithine aminotransferase |
| chr6_-_13783604 | 6.40 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr5_-_26118855 | 6.21 |
ENSDART00000009028
|
ela3l
|
elastase 3 like |
| chr23_+_2778813 | 5.67 |
ENSDART00000142621
|
top1
|
DNA topoisomerase I |
| chr2_+_31475772 | 5.67 |
ENSDART00000130722
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr18_+_46773198 | 5.48 |
ENSDART00000174647
|
CABZ01041495.1
|
|
| chr20_-_25522911 | 5.19 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr13_-_7031033 | 5.12 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr13_+_1369003 | 5.09 |
ENSDART00000146082
|
ifngr1
|
interferon gamma receptor 1 |
| chr17_-_51829310 | 5.02 |
ENSDART00000154544
|
numb
|
numb homolog (Drosophila) |
| chr16_+_52512025 | 4.63 |
ENSDART00000056095
|
fabp10a
|
fatty acid binding protein 10a, liver basic |
| chr14_-_33613794 | 4.57 |
ENSDART00000010022
|
xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr21_+_10739846 | 4.28 |
ENSDART00000084011
|
cplx4a
|
complexin 4a |
| chr9_+_56422311 | 4.27 |
ENSDART00000171958
|
gpr39
|
G protein-coupled receptor 39 |
| chr6_-_9565526 | 4.19 |
ENSDART00000151470
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr7_+_39389273 | 4.09 |
ENSDART00000191298
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr20_-_44576949 | 4.01 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr9_-_1200187 | 4.00 |
ENSDART00000158760
|
ino80da
|
INO80 complex subunit Da |
| chr6_-_35487413 | 4.00 |
ENSDART00000102461
|
rgs8
|
regulator of G protein signaling 8 |
| chr4_-_1324141 | 3.98 |
ENSDART00000180720
|
ptn
|
pleiotrophin |
| chr18_+_45550783 | 3.79 |
ENSDART00000138075
|
kifc3
|
kinesin family member C3 |
| chr19_-_38830582 | 3.72 |
ENSDART00000189966
ENSDART00000183055 |
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr16_-_29334672 | 3.65 |
ENSDART00000162835
|
bcan
|
brevican |
| chr21_+_6212844 | 3.64 |
ENSDART00000150301
|
fnbp1b
|
formin binding protein 1b |
| chr19_-_27261102 | 3.48 |
ENSDART00000143919
|
gabbr1b
|
gamma-aminobutyric acid (GABA) B receptor, 1b |
| chr23_+_20563779 | 3.47 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr6_+_46341306 | 3.44 |
ENSDART00000111905
|
BX649498.1
|
|
| chr2_+_31476065 | 3.44 |
ENSDART00000049219
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr21_-_14630831 | 3.42 |
ENSDART00000132142
ENSDART00000089967 |
cacna1bb
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, b |
| chr25_-_554142 | 3.40 |
ENSDART00000028997
|
myo9ab
|
myosin IXAb |
| chr14_+_40874608 | 3.40 |
ENSDART00000168448
|
si:ch211-106m9.1
|
si:ch211-106m9.1 |
| chr12_-_14922955 | 3.38 |
ENSDART00000002078
|
neurod2
|
neurogenic differentiation 2 |
| chr20_-_39273505 | 3.29 |
ENSDART00000153114
|
clu
|
clusterin |
| chr11_-_28911172 | 3.25 |
ENSDART00000168493
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr1_-_29045426 | 3.21 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr22_-_33679277 | 3.16 |
ENSDART00000169948
|
FO904977.1
|
|
| chr19_-_6134802 | 3.15 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr12_+_31744217 | 3.12 |
ENSDART00000190361
|
RNF157
|
si:dkey-49c17.3 |
| chr3_-_60886984 | 3.09 |
ENSDART00000170974
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr17_-_51202339 | 3.07 |
ENSDART00000167117
|
si:ch1073-469d17.2
|
si:ch1073-469d17.2 |
| chr2_-_37888429 | 3.05 |
ENSDART00000183277
|
mbl2
|
mannose binding lectin 2 |
| chr9_-_22831836 | 3.04 |
ENSDART00000142585
|
neb
|
nebulin |
| chr11_-_37359416 | 3.04 |
ENSDART00000159184
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr21_-_43952958 | 3.04 |
ENSDART00000039571
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr11_-_7050825 | 3.01 |
ENSDART00000179749
|
si:ch211-253b8.5
|
si:ch211-253b8.5 |
| chr23_+_5465806 | 2.93 |
ENSDART00000149434
ENSDART00000148506 |
tulp1a
|
tubby like protein 1a |
| chr6_+_21740672 | 2.88 |
ENSDART00000193734
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr19_+_9174166 | 2.87 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr14_+_15622817 | 2.84 |
ENSDART00000158624
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr10_+_29431529 | 2.82 |
ENSDART00000158154
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr19_-_38872650 | 2.77 |
ENSDART00000146641
|
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr14_-_21219659 | 2.75 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr13_-_30027730 | 2.73 |
ENSDART00000044009
|
scdb
|
stearoyl-CoA desaturase b |
| chr18_+_660578 | 2.72 |
ENSDART00000161203
|
CELF6
|
si:dkey-205h23.2 |
| chr24_-_1657276 | 2.69 |
ENSDART00000168131
|
si:ch73-378g22.1
|
si:ch73-378g22.1 |
| chr14_+_32838110 | 2.67 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr10_-_24362775 | 2.65 |
ENSDART00000182104
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr21_+_13353263 | 2.62 |
ENSDART00000114677
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr1_-_40227166 | 2.60 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr12_-_35949936 | 2.59 |
ENSDART00000192583
|
AL954682.1
|
|
| chr23_-_5032587 | 2.57 |
ENSDART00000163903
|
kcna2b
|
potassium voltage-gated channel, shaker-related subfamily, member 2b |
| chr9_-_9980704 | 2.56 |
ENSDART00000130243
ENSDART00000193475 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr20_-_18731268 | 2.56 |
ENSDART00000183893
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr14_+_32837914 | 2.55 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr8_-_26609259 | 2.54 |
ENSDART00000027301
|
sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr3_+_23092762 | 2.53 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr11_-_32723851 | 2.52 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr21_-_13085242 | 2.52 |
ENSDART00000044504
|
zgc:109965
|
zgc:109965 |
| chr2_+_50626476 | 2.52 |
ENSDART00000018150
|
neurod6b
|
neuronal differentiation 6b |
| chr22_+_17205608 | 2.51 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr13_+_3954540 | 2.49 |
ENSDART00000092646
|
lrrc73
|
leucine rich repeat containing 73 |
| chr5_-_38342992 | 2.48 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr7_+_30823749 | 2.44 |
ENSDART00000085661
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr8_-_3716734 | 2.42 |
ENSDART00000172966
|
bicdl1
|
BICD family like cargo adaptor 1 |
| chr21_-_16632808 | 2.39 |
ENSDART00000172645
|
unc5da
|
unc-5 netrin receptor Da |
| chr20_-_39273987 | 2.37 |
ENSDART00000127173
|
clu
|
clusterin |
| chr6_-_57426097 | 2.36 |
ENSDART00000171531
|
znfx1
|
zinc finger, NFX1-type containing 1 |
| chr6_+_9175886 | 2.36 |
ENSDART00000165333
|
si:ch211-207l14.1
|
si:ch211-207l14.1 |
| chr25_+_7670683 | 2.34 |
ENSDART00000040275
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr24_+_29449690 | 2.32 |
ENSDART00000105743
ENSDART00000193556 ENSDART00000145816 |
ntng1a
|
netrin g1a |
| chr2_+_5948534 | 2.31 |
ENSDART00000124324
ENSDART00000176461 |
slc1a7a
|
solute carrier family 1 (glutamate transporter), member 7a |
| chr1_-_14233815 | 2.29 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr5_+_62611400 | 2.26 |
ENSDART00000132054
|
abr
|
active BCR-related |
| chr8_+_7359294 | 2.25 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr10_-_44508249 | 2.24 |
ENSDART00000160018
|
DUSP26
|
dual specificity phosphatase 26 |
| chr8_-_44868020 | 2.24 |
ENSDART00000142712
|
cacna1fa
|
calcium channel, voltage-dependent, L type, alpha 1F subunit a |
| chr11_+_6159595 | 2.24 |
ENSDART00000178367
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr20_+_1412193 | 2.22 |
ENSDART00000064419
|
leg1.1
|
liver-enriched gene 1, tandem duplicate 1 |
| chr23_+_5470967 | 2.19 |
ENSDART00000110522
ENSDART00000149050 |
tulp1a
|
tubby like protein 1a |
| chr3_+_50069610 | 2.19 |
ENSDART00000056619
|
zgc:103625
|
zgc:103625 |
| chr18_-_38088099 | 2.19 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr21_+_39100289 | 2.18 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr15_+_3284416 | 2.17 |
ENSDART00000187665
ENSDART00000171723 |
foxo1a
|
forkhead box O1 a |
| chr16_+_21738194 | 2.17 |
ENSDART00000163688
|
FP085428.1
|
Danio rerio si:ch211-154o6.4 (si:ch211-154o6.4), mRNA. |
| chr15_+_9053059 | 2.16 |
ENSDART00000012039
|
ppm1na
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Na (putative) |
| chr18_+_9635178 | 2.16 |
ENSDART00000183486
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr18_+_12147971 | 2.15 |
ENSDART00000162067
ENSDART00000168386 |
fgd4a
|
FYVE, RhoGEF and PH domain containing 4a |
| chr17_-_45040813 | 2.15 |
ENSDART00000075514
|
entpd5a
|
ectonucleoside triphosphate diphosphohydrolase 5a |
| chr11_-_28157129 | 2.13 |
ENSDART00000103450
|
lactbl1b
|
lactamase, beta-like 1b |
| chr21_+_27382893 | 2.13 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr15_-_28094256 | 2.12 |
ENSDART00000142041
ENSDART00000132153 ENSDART00000146657 ENSDART00000048720 |
cryba1a
|
crystallin, beta A1a |
| chr5_-_40190949 | 2.12 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr8_-_49935358 | 2.12 |
ENSDART00000159782
ENSDART00000156841 |
agtpbp1
CU694202.1
|
ATP/GTP binding protein 1 |
| chr6_-_27139396 | 2.10 |
ENSDART00000055848
|
zgc:103559
|
zgc:103559 |
| chr24_-_17444067 | 2.08 |
ENSDART00000155843
|
cntnap2a
|
contactin associated protein like 2a |
| chr7_+_10351038 | 2.06 |
ENSDART00000173256
|
si:cabz01029535.1
|
si:cabz01029535.1 |
| chr21_+_8427059 | 2.06 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr13_-_44285793 | 2.05 |
ENSDART00000167383
|
CABZ01069436.1
|
|
| chr15_+_30158652 | 2.05 |
ENSDART00000190682
|
nlk2
|
nemo-like kinase, type 2 |
| chr6_+_22597362 | 2.04 |
ENSDART00000131242
|
cygb2
|
cytoglobin 2 |
| chr13_+_228045 | 2.04 |
ENSDART00000161091
|
zgc:64201
|
zgc:64201 |
| chr18_-_38087875 | 2.04 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr6_+_54538948 | 2.04 |
ENSDART00000149270
|
tulp1b
|
tubby like protein 1b |
| chr5_+_21891305 | 2.03 |
ENSDART00000136788
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr5_-_18513950 | 2.02 |
ENSDART00000145878
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr25_+_4541211 | 2.02 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr5_-_23280098 | 2.01 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr11_-_43226255 | 2.00 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr10_-_17103651 | 1.98 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr25_-_16818978 | 1.98 |
ENSDART00000104140
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr16_-_31644545 | 1.94 |
ENSDART00000181634
|
CR855311.5
|
|
| chr6_-_43449013 | 1.92 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr15_+_42933236 | 1.92 |
ENSDART00000167763
|
slc8a2b
|
solute carrier family 8 (sodium/calcium exchanger), member 2b |
| chr3_+_26144765 | 1.92 |
ENSDART00000146267
ENSDART00000043932 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr20_-_25669813 | 1.91 |
ENSDART00000153118
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr14_-_4556896 | 1.91 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr15_-_29598679 | 1.88 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr5_+_69950882 | 1.88 |
ENSDART00000097359
|
dnajc25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr5_-_51819027 | 1.88 |
ENSDART00000164267
|
homer1b
|
homer scaffolding protein 1b |
| chr7_+_32722227 | 1.88 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr9_+_31795343 | 1.86 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr25_-_20258508 | 1.85 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr7_+_26224211 | 1.85 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr10_-_5847904 | 1.85 |
ENSDART00000161096
|
ankrd55
|
ankyrin repeat domain 55 |
| chr23_-_1660708 | 1.84 |
ENSDART00000175138
|
CU693481.1
|
|
| chr19_-_10373630 | 1.84 |
ENSDART00000131517
|
si:ch211-232m10.6
|
si:ch211-232m10.6 |
| chr3_+_50312422 | 1.84 |
ENSDART00000157689
|
gas7a
|
growth arrest-specific 7a |
| chr22_-_21897203 | 1.82 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr9_+_25096500 | 1.82 |
ENSDART00000135074
ENSDART00000180436 ENSDART00000108629 |
lrch1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr19_+_43669122 | 1.78 |
ENSDART00000139151
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr17_+_32360673 | 1.78 |
ENSDART00000155519
|
si:ch211-139d20.3
|
si:ch211-139d20.3 |
| chr8_+_8735285 | 1.78 |
ENSDART00000165655
|
elk1
|
ELK1, member of ETS oncogene family |
| chr7_+_29133321 | 1.78 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr21_-_22737228 | 1.77 |
ENSDART00000151366
|
fbxo40.2
|
F-box protein 40, tandem duplicate 2 |
| chr1_-_22370660 | 1.77 |
ENSDART00000127506
|
si:ch73-380n15.2
|
si:ch73-380n15.2 |
| chr17_-_25397558 | 1.76 |
ENSDART00000182052
|
fam167b
|
family with sequence similarity 167, member B |
| chr9_+_56422517 | 1.76 |
ENSDART00000168620
|
gpr39
|
G protein-coupled receptor 39 |
| chr1_+_21937201 | 1.76 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr3_+_33340939 | 1.74 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr5_-_33769211 | 1.74 |
ENSDART00000133504
|
dab2ipb
|
DAB2 interacting protein b |
| chr6_-_51101834 | 1.73 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr3_+_50310684 | 1.73 |
ENSDART00000112152
|
gas7a
|
growth arrest-specific 7a |
| chr13_+_28854438 | 1.72 |
ENSDART00000193407
ENSDART00000189554 |
CU639469.1
|
|
| chr13_+_3954715 | 1.71 |
ENSDART00000182477
ENSDART00000192142 ENSDART00000190962 |
lrrc73
|
leucine rich repeat containing 73 |
| chr10_-_24371312 | 1.71 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr5_+_3501859 | 1.71 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr13_-_27916439 | 1.71 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr10_-_29120515 | 1.70 |
ENSDART00000162016
ENSDART00000149140 |
zpld1a
|
zona pellucida-like domain containing 1a |
| chr8_+_36666789 | 1.70 |
ENSDART00000144255
|
magixb
|
MAGI family member, X-linked b |
| chr6_-_35401282 | 1.69 |
ENSDART00000127612
|
rgs5a
|
regulator of G protein signaling 5a |
| chr7_-_28463106 | 1.69 |
ENSDART00000137799
|
trim66
|
tripartite motif containing 66 |
| chr6_+_39493864 | 1.67 |
ENSDART00000086263
|
mettl7a
|
methyltransferase like 7A |
| chr17_-_44584811 | 1.67 |
ENSDART00000165059
ENSDART00000165252 |
slc35f4
|
solute carrier family 35, member F4 |
| chr23_+_21544227 | 1.64 |
ENSDART00000140253
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr24_-_38644937 | 1.64 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr4_-_20181964 | 1.62 |
ENSDART00000022539
|
fgl2a
|
fibrinogen-like 2a |
| chr18_+_10840071 | 1.61 |
ENSDART00000014496
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr24_+_32472155 | 1.59 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr12_+_21525496 | 1.58 |
ENSDART00000152974
|
ca10a
|
carbonic anhydrase Xa |
| chr2_+_33541928 | 1.57 |
ENSDART00000162852
|
BX548164.1
|
|
| chr25_+_33849647 | 1.56 |
ENSDART00000121449
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr14_-_21218891 | 1.55 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr14_+_40641350 | 1.54 |
ENSDART00000074506
|
tnmd
|
tenomodulin |
| chr13_-_40499296 | 1.54 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr19_+_43684376 | 1.54 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr11_+_6902946 | 1.53 |
ENSDART00000144006
|
crtc1b
|
CREB regulated transcription coactivator 1b |
| chr25_-_16818380 | 1.53 |
ENSDART00000155401
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr17_-_52579709 | 1.52 |
ENSDART00000156806
|
rps6kl1
|
ribosomal protein S6 kinase-like 1 |
| chr9_+_49712868 | 1.49 |
ENSDART00000192969
ENSDART00000183310 |
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr9_+_28693592 | 1.49 |
ENSDART00000110198
|
zgc:162780
|
zgc:162780 |
| chr18_+_48576527 | 1.49 |
ENSDART00000167555
|
kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr2_+_30878864 | 1.48 |
ENSDART00000009326
|
oprk1
|
opioid receptor, kappa 1 |
| chr18_+_3634652 | 1.48 |
ENSDART00000159913
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr18_+_15795748 | 1.46 |
ENSDART00000147024
ENSDART00000137681 |
nudt4a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4a |
| chr3_+_22242269 | 1.45 |
ENSDART00000168970
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr1_+_32521469 | 1.45 |
ENSDART00000113818
ENSDART00000152580 |
nlgn4a
|
neuroligin 4a |
| chr13_-_16226312 | 1.44 |
ENSDART00000163952
|
zgc:110045
|
zgc:110045 |
| chr7_-_23996133 | 1.44 |
ENSDART00000173761
|
si:dkey-183c6.8
|
si:dkey-183c6.8 |
| chr6_-_46565466 | 1.43 |
ENSDART00000153673
|
kcnb1
|
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr6_-_35032792 | 1.42 |
ENSDART00000168256
|
ddr2a
|
discoidin domain receptor tyrosine kinase 2a |
| chr5_+_37854685 | 1.42 |
ENSDART00000051222
ENSDART00000185283 |
ins
|
preproinsulin |
| chr2_-_37478418 | 1.41 |
ENSDART00000146103
|
dapk3
|
death-associated protein kinase 3 |
| chr11_+_29999414 | 1.40 |
ENSDART00000161160
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr5_-_52243424 | 1.40 |
ENSDART00000159078
|
erap2
|
endoplasmic reticulum aminopeptidase 2 |
| chr19_-_9882821 | 1.39 |
ENSDART00000147128
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr5_+_20147830 | 1.39 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
Network of associatons between targets according to the STRING database.
First level regulatory network of neurod1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 1.1 | 3.2 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.9 | 3.5 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.8 | 3.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.7 | 2.9 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.7 | 6.5 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.7 | 2.1 | GO:0000256 | allantoin catabolic process(GO:0000256) purine nucleobase catabolic process(GO:0006145) |
| 0.7 | 2.7 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.7 | 3.4 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.6 | 7.0 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.6 | 1.8 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.6 | 2.4 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.6 | 2.8 | GO:1901908 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.5 | 2.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.5 | 3.0 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.5 | 2.0 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.5 | 1.9 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.4 | 3.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.4 | 5.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.4 | 4.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.4 | 2.1 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.4 | 3.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.4 | 1.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.4 | 1.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.4 | 1.1 | GO:1901546 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.4 | 0.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.4 | 1.9 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.4 | 1.5 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.4 | 1.4 | GO:0048521 | positive regulation of epithelial cell differentiation(GO:0030858) negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.3 | 2.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.3 | 1.0 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.3 | 5.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.3 | 3.3 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.3 | 2.0 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.3 | 0.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.3 | 1.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.3 | 1.9 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.3 | 0.8 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 9.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 4.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.2 | 4.0 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.2 | 1.2 | GO:0060402 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.2 | 1.8 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 2.2 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.2 | 2.2 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.2 | 2.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.2 | 2.2 | GO:1990709 | presynaptic active zone assembly(GO:1904071) presynaptic active zone organization(GO:1990709) |
| 0.2 | 2.1 | GO:0055062 | phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.2 | 1.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 0.6 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.2 | 0.8 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 1.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 0.6 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.2 | 6.5 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.2 | 5.9 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.2 | 0.8 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.2 | 2.7 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.2 | 0.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.7 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 1.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 6.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 6.5 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.4 | GO:0060262 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.1 | 2.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 2.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.4 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 1.4 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 1.2 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.9 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 1.7 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 1.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.3 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 2.0 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 5.2 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.0 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 2.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.8 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.1 | 1.9 | GO:0098703 | inorganic cation import into cell(GO:0098659) calcium ion import across plasma membrane(GO:0098703) inorganic ion import into cell(GO:0099587) calcium ion import into cell(GO:1990035) |
| 0.1 | 4.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.9 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 1.8 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.6 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 5.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.9 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.1 | 2.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 1.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 2.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 2.2 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 2.5 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.1 | 0.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 2.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 1.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 1.4 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 0.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 5.8 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 1.7 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 1.0 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 2.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 1.2 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 0.5 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 0.5 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.7 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.1 | 2.9 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 5.0 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.1 | 4.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 2.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 2.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 1.4 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 1.9 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 7.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.4 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.2 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.9 | GO:0050927 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.1 | 1.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 4.1 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.6 | GO:0051928 | positive regulation of calcium ion transport(GO:0051928) |
| 0.0 | 0.3 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 3.0 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 1.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.9 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.3 | GO:0090311 | regulation of histone deacetylation(GO:0031063) regulation of protein deacetylation(GO:0090311) |
| 0.0 | 0.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.1 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 2.5 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 1.3 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 2.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 4.7 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 1.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.9 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 4.1 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.9 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 1.1 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.9 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.9 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.4 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.4 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 0.6 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.3 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.3 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 2.6 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.6 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.4 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 1.3 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.5 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 1.4 | GO:0010942 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.5 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.6 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 7.3 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 0.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.4 | GO:0032944 | regulation of mononuclear cell proliferation(GO:0032944) regulation of T cell proliferation(GO:0042129) regulation of lymphocyte proliferation(GO:0050670) regulation of leukocyte proliferation(GO:0070663) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.6 | 5.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.5 | 1.9 | GO:0031673 | H zone(GO:0031673) |
| 0.4 | 3.5 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.4 | 2.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 1.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.3 | 2.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 6.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 8.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 2.2 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.2 | 2.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 14.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 3.3 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.2 | 1.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 2.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 2.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 1.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.4 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.7 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 2.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 3.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.3 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 4.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.7 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.3 | GO:0044297 | cell body(GO:0044297) |
| 0.1 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 6.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 3.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 4.4 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.4 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.4 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 1.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 3.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 4.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.3 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 2.7 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 2.2 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.0 | 1.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.1 | GO:0030880 | RNA polymerase complex(GO:0030880) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.6 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 19.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.1 | GO:0098794 | postsynapse(GO:0098794) |
| 0.0 | 6.1 | GO:0097458 | neuron part(GO:0097458) |
| 0.0 | 1.3 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.7 | 2.7 | GO:0032896 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.6 | 4.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.6 | 1.8 | GO:0042166 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.6 | 4.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.6 | 2.8 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.5 | 2.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.4 | 5.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.4 | 3.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.4 | 11.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.4 | 1.9 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.4 | 1.8 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 3.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.3 | 1.7 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.3 | 1.4 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.3 | 2.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 5.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.3 | 2.0 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.3 | 1.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.3 | 2.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 1.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 0.8 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.3 | 1.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 1.2 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.2 | 0.7 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 2.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 0.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 1.9 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.2 | 0.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 1.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 2.0 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 1.8 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 9.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 0.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 0.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 0.7 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 6.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 2.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 1.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.2 | 2.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 1.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 3.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 2.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.3 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 3.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 2.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 2.0 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 9.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 3.8 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.2 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.7 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.1 | 5.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 2.6 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 6.5 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 0.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.6 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.4 | GO:0047611 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.1 | 1.4 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.9 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.8 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 0.8 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 4.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 3.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.1 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.3 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.1 | 1.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 5.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 2.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.8 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 1.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 3.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 6.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 0.7 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.5 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 1.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 0.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 1.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 4.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 12.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.0 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.4 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 5.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 17.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.5 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 1.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 1.0 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 1.1 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 5.6 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 2.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.4 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 16.2 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 5.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.7 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 1.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 6.6 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 7.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 3.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 5.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 4.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 6.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.7 | 5.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.6 | 6.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 2.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 3.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 3.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 3.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 5.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 1.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 3.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 4.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 5.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 0.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 5.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.9 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.1 | 1.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 2.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 4.3 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 2.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |