Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
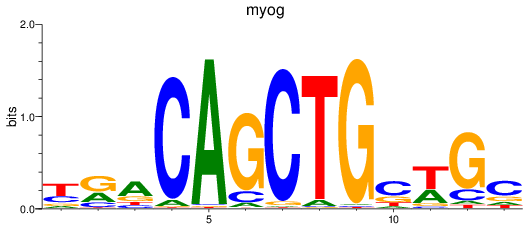
Results for myog
Z-value: 0.86
Transcription factors associated with myog
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myog
|
ENSDARG00000009438 | myogenin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myog | dr11_v1_chr11_-_22599584_22599584 | 0.71 | 6.5e-04 | Click! |
Activity profile of myog motif
Sorted Z-values of myog motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_46201008 | 5.11 |
ENSDART00000160110
|
tgm1l4
|
transglutaminase 1 like 4 |
| chr21_-_217589 | 2.31 |
ENSDART00000185017
|
CZQB01146713.1
|
|
| chr17_-_14815557 | 2.13 |
ENSDART00000154473
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr7_+_2455344 | 1.86 |
ENSDART00000172942
|
si:dkey-125e8.4
|
si:dkey-125e8.4 |
| chr20_+_43691208 | 1.78 |
ENSDART00000152976
ENSDART00000045185 |
lin9
|
lin-9 DREAM MuvB core complex component |
| chr19_-_47571456 | 1.73 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr20_-_29498178 | 1.66 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr7_+_44715224 | 1.65 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr7_+_49715750 | 1.61 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr21_-_19919020 | 1.60 |
ENSDART00000147396
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr19_+_15440841 | 1.57 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr5_+_32835219 | 1.56 |
ENSDART00000140832
ENSDART00000186055 |
si:ch211-208h16.4
|
si:ch211-208h16.4 |
| chr17_+_132555 | 1.56 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr19_+_15441022 | 1.53 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr5_-_63515210 | 1.40 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr17_-_28097760 | 1.40 |
ENSDART00000149861
|
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr11_-_18015534 | 1.39 |
ENSDART00000181953
|
qrich1
|
glutamine-rich 1 |
| chr14_-_25928899 | 1.37 |
ENSDART00000143518
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr25_+_3294150 | 1.33 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr23_+_44461493 | 1.32 |
ENSDART00000149854
|
si:ch1073-228j22.1
|
si:ch1073-228j22.1 |
| chr2_-_11027258 | 1.30 |
ENSDART00000081072
ENSDART00000193824 ENSDART00000187036 ENSDART00000097741 |
ssbp3a
|
single stranded DNA binding protein 3a |
| chr8_-_18613948 | 1.28 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr13_+_30903816 | 1.26 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr24_+_20575259 | 1.25 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr17_-_14836320 | 1.22 |
ENSDART00000157051
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr16_+_19537073 | 1.21 |
ENSDART00000190590
|
sp8b
|
sp8 transcription factor b |
| chr19_-_30810328 | 1.20 |
ENSDART00000184875
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr11_-_41966854 | 1.20 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr6_+_40629066 | 1.19 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr16_+_19536614 | 1.18 |
ENSDART00000112894
ENSDART00000079201 ENSDART00000139357 |
sp8b
|
sp8 transcription factor b |
| chr9_+_21306902 | 1.17 |
ENSDART00000138554
ENSDART00000004108 |
xpo4
|
exportin 4 |
| chr2_-_17114852 | 1.17 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr6_-_39313027 | 1.15 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr13_+_11439486 | 1.14 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr14_-_26482096 | 1.14 |
ENSDART00000187280
|
smad5
|
SMAD family member 5 |
| chr4_-_52165969 | 1.13 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr15_-_39969988 | 1.13 |
ENSDART00000146054
|
rps5
|
ribosomal protein S5 |
| chr3_-_29962345 | 1.12 |
ENSDART00000136819
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr1_+_23563691 | 1.12 |
ENSDART00000142879
|
ncapg
|
non-SMC condensin I complex, subunit G |
| chr3_-_19368435 | 1.11 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr5_-_14509137 | 1.11 |
ENSDART00000180742
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr5_-_8164439 | 1.11 |
ENSDART00000189912
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr16_-_42894628 | 1.09 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr3_+_59935606 | 1.08 |
ENSDART00000154157
|
arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr20_+_34502606 | 1.05 |
ENSDART00000139739
|
gorab
|
golgin, rab6-interacting |
| chr17_+_1360192 | 1.04 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr24_+_25210015 | 1.03 |
ENSDART00000081043
|
cip2a
|
cell proliferation regulating inhibitor of protein phosphatase 2A |
| chr6_-_32093830 | 1.03 |
ENSDART00000017695
|
foxd3
|
forkhead box D3 |
| chr23_-_19831739 | 1.00 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr10_+_33588715 | 0.99 |
ENSDART00000051198
|
mis18a
|
MIS18 kinetochore protein A |
| chr4_-_76488581 | 0.98 |
ENSDART00000174291
|
ftr51
|
finTRIM family, member 51 |
| chr22_-_10541372 | 0.97 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr13_+_24263049 | 0.97 |
ENSDART00000135992
ENSDART00000088005 |
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr2_-_21335131 | 0.97 |
ENSDART00000057022
|
klhl40a
|
kelch-like family member 40a |
| chr9_+_41024973 | 0.97 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr19_-_47570672 | 0.96 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr13_+_51579851 | 0.96 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr1_+_38142354 | 0.96 |
ENSDART00000179352
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr2_-_17115256 | 0.95 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr19_+_18812805 | 0.95 |
ENSDART00000187164
ENSDART00000172412 |
cratb
|
carnitine O-acetyltransferase b |
| chr16_+_26706519 | 0.93 |
ENSDART00000142706
|
virma
|
vir like m6A methyltransferase associated |
| chr14_-_41388178 | 0.93 |
ENSDART00000124532
ENSDART00000125016 ENSDART00000169247 |
cstf2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2 |
| chr8_-_11229523 | 0.93 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr8_-_1051438 | 0.93 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr25_-_14637660 | 0.92 |
ENSDART00000143666
|
nav2b
|
neuron navigator 2b |
| chr5_+_17780475 | 0.92 |
ENSDART00000110783
ENSDART00000115227 |
chfr
|
checkpoint with forkhead and ring finger domains, E3 ubiquitin protein ligase |
| chr11_+_36477481 | 0.92 |
ENSDART00000128245
|
ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr2_+_16173999 | 0.91 |
ENSDART00000177639
ENSDART00000157601 ENSDART00000190186 ENSDART00000045933 |
sh3glb1b
|
SH3-domain GRB2-like endophilin B1b |
| chr8_-_50259448 | 0.90 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr13_+_2894536 | 0.89 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr18_-_20444296 | 0.89 |
ENSDART00000132993
|
kif23
|
kinesin family member 23 |
| chr10_-_44560165 | 0.88 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr9_+_32872690 | 0.88 |
ENSDART00000020798
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr8_-_14609284 | 0.88 |
ENSDART00000146175
|
cep350
|
centrosomal protein 350 |
| chr16_-_6205790 | 0.87 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr5_+_31779911 | 0.87 |
ENSDART00000098163
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr4_+_8638622 | 0.87 |
ENSDART00000186829
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr15_-_2652640 | 0.86 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr8_-_37043900 | 0.85 |
ENSDART00000139567
|
renbp
|
renin binding protein |
| chr5_+_250078 | 0.85 |
ENSDART00000127504
|
trabd2a
|
TraB domain containing 2A |
| chr12_+_6002715 | 0.84 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr8_+_39570615 | 0.84 |
ENSDART00000142557
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr18_+_30441740 | 0.83 |
ENSDART00000189074
|
gse1
|
Gse1 coiled-coil protein |
| chr4_+_10066840 | 0.83 |
ENSDART00000026492
|
flncb
|
filamin C, gamma b (actin binding protein 280) |
| chr13_-_13030851 | 0.82 |
ENSDART00000009499
|
nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr24_-_9300160 | 0.82 |
ENSDART00000152378
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr23_-_19977214 | 0.82 |
ENSDART00000054659
|
ccnq
|
cyclin Q |
| chr7_+_41265618 | 0.82 |
ENSDART00000173688
|
scrib
|
scribbled planar cell polarity protein |
| chr17_-_51818659 | 0.82 |
ENSDART00000111389
ENSDART00000157244 |
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr2_+_32016516 | 0.81 |
ENSDART00000135040
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr20_+_1316495 | 0.81 |
ENSDART00000064439
|
nup43
|
nucleoporin 43 |
| chr9_-_29578037 | 0.81 |
ENSDART00000189026
|
cenpj
|
centromere protein J |
| chr11_-_17755444 | 0.80 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr14_-_9281232 | 0.80 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr16_-_31469065 | 0.80 |
ENSDART00000182397
|
si:ch211-251p5.5
|
si:ch211-251p5.5 |
| chr17_+_27434626 | 0.79 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr8_-_43677762 | 0.79 |
ENSDART00000167762
|
ep400
|
E1A binding protein p400 |
| chr23_+_20931030 | 0.78 |
ENSDART00000167014
|
pax7b
|
paired box 7b |
| chr19_+_26072624 | 0.78 |
ENSDART00000147627
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr16_+_33953644 | 0.77 |
ENSDART00000164447
ENSDART00000159969 |
arid1aa
|
AT rich interactive domain 1Aa (SWI-like) |
| chr14_+_7939398 | 0.77 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr12_+_47663419 | 0.77 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr16_-_54810464 | 0.77 |
ENSDART00000030658
|
zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr18_-_19819812 | 0.76 |
ENSDART00000060344
|
aagab
|
alpha and gamma adaptin binding protein |
| chr17_-_10073926 | 0.76 |
ENSDART00000166081
ENSDART00000161574 |
zgc:109986
|
zgc:109986 |
| chr14_+_7939216 | 0.76 |
ENSDART00000171657
|
cxxc5b
|
CXXC finger protein 5b |
| chr5_-_19400166 | 0.76 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr4_+_77993977 | 0.75 |
ENSDART00000174118
|
terfa
|
telomeric repeat binding factor a |
| chr14_+_28486213 | 0.75 |
ENSDART00000161852
|
stag2b
|
stromal antigen 2b |
| chr25_+_28589669 | 0.75 |
ENSDART00000193901
|
tmem17
|
transmembrane protein 17 |
| chr12_+_48681601 | 0.74 |
ENSDART00000187831
|
uros
|
uroporphyrinogen III synthase |
| chr5_-_20135679 | 0.74 |
ENSDART00000079402
|
usp30
|
ubiquitin specific peptidase 30 |
| chr7_-_41726657 | 0.73 |
ENSDART00000099121
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr22_+_465269 | 0.72 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr19_-_46957968 | 0.72 |
ENSDART00000043713
|
angpt1
|
angiopoietin 1 |
| chr17_+_24597001 | 0.72 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr19_-_24757231 | 0.72 |
ENSDART00000128177
|
si:dkey-154b15.1
|
si:dkey-154b15.1 |
| chr22_+_11153590 | 0.71 |
ENSDART00000188207
|
bcor
|
BCL6 corepressor |
| chr23_+_553396 | 0.70 |
ENSDART00000034707
|
lsm14b
|
LSM family member 14B |
| chr7_-_38570299 | 0.70 |
ENSDART00000143815
|
celf1
|
cugbp, Elav-like family member 1 |
| chr25_-_25619550 | 0.70 |
ENSDART00000150400
|
tbc1d2b
|
TBC1 domain family, member 2B |
| chr11_-_287670 | 0.69 |
ENSDART00000035737
|
slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr3_+_46559639 | 0.69 |
ENSDART00000146189
ENSDART00000127832 ENSDART00000151035 |
raver1
|
ribonucleoprotein, PTB-binding 1 |
| chr4_-_76488854 | 0.69 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr8_-_4573545 | 0.69 |
ENSDART00000127153
|
dhx37
|
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
| chr7_+_5911670 | 0.68 |
ENSDART00000173025
|
si:dkey-23a13.9
|
si:dkey-23a13.9 |
| chr13_+_25286538 | 0.68 |
ENSDART00000180282
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr6_+_59991076 | 0.68 |
ENSDART00000163575
|
CABZ01100888.1
|
|
| chr25_-_14433503 | 0.68 |
ENSDART00000103957
|
exoc3l1
|
exocyst complex component 3-like 1 |
| chr14_+_10596628 | 0.67 |
ENSDART00000115177
|
gpr174
|
G protein-coupled receptor 174 |
| chr11_+_6136220 | 0.67 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr21_-_30026359 | 0.67 |
ENSDART00000153645
|
pwwp2a
|
PWWP domain containing 2A |
| chr14_+_21755469 | 0.67 |
ENSDART00000186326
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr14_-_22118055 | 0.66 |
ENSDART00000179892
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr4_+_13412030 | 0.66 |
ENSDART00000003694
|
cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr1_+_41609676 | 0.66 |
ENSDART00000183675
|
mogs
|
mannosyl-oligosaccharide glucosidase |
| chr19_-_2317558 | 0.65 |
ENSDART00000190300
|
sp8a
|
sp8 transcription factor a |
| chr4_-_59152337 | 0.65 |
ENSDART00000156770
|
znf1149
|
zinc finger protein 1149 |
| chr9_+_22657221 | 0.65 |
ENSDART00000101765
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr12_+_49125510 | 0.65 |
ENSDART00000185804
|
FO704607.1
|
|
| chr13_-_31622195 | 0.64 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr24_+_39108243 | 0.64 |
ENSDART00000156353
|
mss51
|
MSS51 mitochondrial translational activator |
| chr1_-_20271138 | 0.64 |
ENSDART00000185931
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr14_-_7375049 | 0.64 |
ENSDART00000054809
|
trappc11
|
trafficking protein particle complex 11 |
| chr23_-_3408777 | 0.64 |
ENSDART00000193245
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr4_-_21466480 | 0.63 |
ENSDART00000139962
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr2_-_22492289 | 0.63 |
ENSDART00000168022
|
gtf2b
|
general transcription factor IIB |
| chr24_-_17389263 | 0.63 |
ENSDART00000122757
|
cul1b
|
cullin 1b |
| chr6_-_32703317 | 0.63 |
ENSDART00000064833
|
mafaa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
| chr3_-_15139293 | 0.62 |
ENSDART00000161213
ENSDART00000146053 |
fam173a
|
family with sequence similarity 173, member A |
| chr17_+_53296851 | 0.62 |
ENSDART00000158313
|
ddx24
|
DEAD (Asp-Glu-Ala-Asp) box helicase 24 |
| chr2_-_24402341 | 0.61 |
ENSDART00000155442
ENSDART00000088572 |
zgc:154006
|
zgc:154006 |
| chr13_+_43639867 | 0.61 |
ENSDART00000042588
ENSDART00000074728 |
zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr14_-_30050 | 0.61 |
ENSDART00000164411
|
zbtb49
|
zinc finger and BTB domain containing 49 |
| chr15_-_4568154 | 0.61 |
ENSDART00000155254
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr12_-_10476448 | 0.60 |
ENSDART00000106172
|
rac1a
|
Rac family small GTPase 1a |
| chr15_+_21254800 | 0.60 |
ENSDART00000142902
|
usf1
|
upstream transcription factor 1 |
| chr17_+_23968214 | 0.59 |
ENSDART00000183053
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr2_+_6253246 | 0.59 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr7_+_37742299 | 0.59 |
ENSDART00000143300
|
brd7
|
bromodomain containing 7 |
| chr24_+_26337623 | 0.58 |
ENSDART00000145637
|
mynn
|
myoneurin |
| chr4_+_8532580 | 0.58 |
ENSDART00000162065
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr20_-_33497128 | 0.58 |
ENSDART00000101974
|
erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr13_-_3155243 | 0.58 |
ENSDART00000139183
ENSDART00000050934 |
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr9_-_32300783 | 0.58 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr14_+_21828993 | 0.58 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr12_+_15165736 | 0.57 |
ENSDART00000180398
|
adprh
|
ADP-ribosylarginine hydrolase |
| chr8_-_1255321 | 0.57 |
ENSDART00000149605
|
cdc14b
|
cell division cycle 14B |
| chr2_+_29995590 | 0.57 |
ENSDART00000151906
|
rbm33b
|
RNA binding motif protein 33b |
| chr19_-_43657468 | 0.57 |
ENSDART00000150940
|
si:ch211-193k19.2
|
si:ch211-193k19.2 |
| chr20_+_46268906 | 0.56 |
ENSDART00000113372
|
taar14e
|
trace amine associated receptor 14e |
| chr7_-_20582842 | 0.56 |
ENSDART00000169750
ENSDART00000111719 |
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr14_-_5817039 | 0.55 |
ENSDART00000131820
|
kazald2
|
Kazal-type serine peptidase inhibitor domain 2 |
| chr9_-_105135 | 0.55 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr3_-_8285123 | 0.55 |
ENSDART00000158699
ENSDART00000138588 |
trim35-9
|
tripartite motif containing 35-9 |
| chr11_-_10058 | 0.54 |
ENSDART00000173070
|
myg1
|
melanocyte proliferating gene 1 |
| chr12_-_26415499 | 0.54 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr9_-_32300611 | 0.54 |
ENSDART00000127938
|
hspd1
|
heat shock 60 protein 1 |
| chr23_+_27789795 | 0.54 |
ENSDART00000141458
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr21_-_22317920 | 0.54 |
ENSDART00000191083
ENSDART00000108701 |
gdpd4b
|
glycerophosphodiester phosphodiesterase domain containing 4b |
| chr23_+_16948049 | 0.54 |
ENSDART00000139449
|
si:dkey-147f3.4
|
si:dkey-147f3.4 |
| chr10_+_15454745 | 0.54 |
ENSDART00000129441
ENSDART00000123935 ENSDART00000163446 ENSDART00000087680 ENSDART00000193752 |
erbin
|
erbb2 interacting protein |
| chr19_+_6910384 | 0.54 |
ENSDART00000151389
|
si:ch73-367f21.4
|
si:ch73-367f21.4 |
| chr2_+_36620011 | 0.53 |
ENSDART00000177428
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr4_+_18442941 | 0.53 |
ENSDART00000190819
|
ncaph2
|
non-SMC condensin II complex, subunit H2 |
| chr10_-_25328814 | 0.53 |
ENSDART00000123820
|
tmem135
|
transmembrane protein 135 |
| chr10_-_22803740 | 0.53 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr20_+_25904199 | 0.53 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr20_+_23625387 | 0.53 |
ENSDART00000147945
ENSDART00000150497 |
palld
|
palladin, cytoskeletal associated protein |
| chr21_+_33249478 | 0.53 |
ENSDART00000169972
|
si:ch211-151g22.1
|
si:ch211-151g22.1 |
| chr23_+_26079467 | 0.52 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr20_+_19066858 | 0.52 |
ENSDART00000192086
|
sox7
|
SRY (sex determining region Y)-box 7 |
| chr3_-_49163683 | 0.51 |
ENSDART00000166146
|
dnajb1a
|
DnaJ (Hsp40) homolog, subfamily B, member 1a |
| chr7_-_41468942 | 0.51 |
ENSDART00000174087
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr25_-_16076257 | 0.51 |
ENSDART00000140780
|
ovch2
|
ovochymase 2 |
| chr10_+_589501 | 0.51 |
ENSDART00000188415
|
LO018557.1
|
|
| chr9_-_12885201 | 0.50 |
ENSDART00000124957
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr19_+_14454306 | 0.50 |
ENSDART00000161965
|
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr18_+_6857071 | 0.50 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr23_-_7594723 | 0.50 |
ENSDART00000115298
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of myog
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.7 | 2.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.5 | 1.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.5 | 2.4 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.4 | 4.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 0.9 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.3 | 1.4 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.3 | 1.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.3 | 1.1 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.3 | 1.1 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.3 | 0.8 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.2 | 1.0 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 5.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 0.7 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 0.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 1.0 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.2 | 1.2 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.2 | 1.4 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 0.8 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.2 | 0.7 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.2 | 0.7 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 1.3 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 2.1 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 0.7 | GO:0061113 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.2 | 0.7 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.2 | 1.0 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 0.6 | GO:0014857 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.2 | 0.8 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.2 | 0.5 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.4 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.8 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.1 | 0.7 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.8 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.9 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.8 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 2.0 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.5 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 0.8 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 0.5 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 1.2 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.1 | 1.0 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.3 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.6 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.1 | 0.4 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.4 | GO:1904355 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.1 | 1.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.5 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 0.9 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.3 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.1 | 0.4 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 1.1 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.3 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.6 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.9 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.4 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 0.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.4 | GO:0016037 | light absorption(GO:0016037) |
| 0.1 | 3.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.7 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.4 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 0.7 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.9 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 1.1 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.1 | 0.2 | GO:0060306 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 | 0.3 | GO:0044806 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 0.3 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.3 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.6 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.7 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.2 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.6 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 1.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.5 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.4 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.7 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.1 | 0.5 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 1.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.2 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.6 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 1.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.4 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.4 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 1.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.6 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.2 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.2 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 0.4 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.0 | 0.5 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.5 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:1903573 | IRE1-mediated unfolded protein response(GO:0036498) negative regulation of response to endoplasmic reticulum stress(GO:1903573) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.0 | 0.9 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.6 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.4 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.1 | GO:0021695 | cerebellar Purkinje cell layer development(GO:0021680) cerebellar cortex development(GO:0021695) |
| 0.0 | 0.7 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.4 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 1.1 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.3 | GO:1903076 | regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.0 | 0.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 1.5 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.9 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.2 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.2 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.0 | 1.2 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.6 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 1.0 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.8 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.3 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.6 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.1 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 1.4 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.5 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.6 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 1.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 1.2 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.4 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.0 | 0.0 | GO:1903817 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 1.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.7 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.0 | 0.8 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.7 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.1 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.7 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.3 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.4 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.5 | GO:0006402 | mRNA catabolic process(GO:0006402) |
| 0.0 | 0.6 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 1.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 1.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 0.7 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 4.1 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.0 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.8 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 1.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.6 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.5 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.1 | 1.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 2.4 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 0.8 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.4 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.5 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 3.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 1.3 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 1.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.7 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 2.2 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 2.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.0 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.0 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.4 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.5 | 1.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.4 | 2.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.3 | 1.1 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.3 | 1.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 5.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 0.7 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.2 | 0.8 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 1.4 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.2 | 0.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 1.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 0.7 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.4 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.1 | 0.4 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 1.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 1.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.8 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 1.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.4 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.4 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.5 | GO:1990931 | RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.1 | 0.8 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 1.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.3 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.6 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.1 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.8 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.5 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.3 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 1.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.4 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.1 | 0.6 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.8 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 1.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.6 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 1.0 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.9 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.2 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.5 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.6 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 3.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.3 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.0 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.8 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 1.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.9 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.4 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 2.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.4 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 3.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 4.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 4.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 2.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.4 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 0.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.3 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 1.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.7 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 1.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |