Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
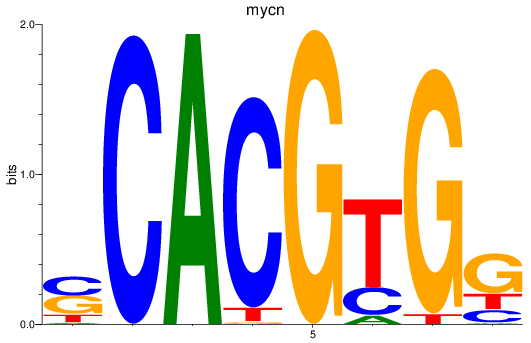
Results for mycn
Z-value: 1.74
Transcription factors associated with mycn
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mycn
|
ENSDARG00000006837 | MYCN proto-oncogene, bHLH transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mycn | dr11_v1_chr20_+_33294428_33294428 | 0.67 | 1.8e-03 | Click! |
Activity profile of mycn motif
Sorted Z-values of mycn motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_25626198 | 6.47 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr20_-_25626428 | 6.19 |
ENSDART00000136475
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr23_+_32101361 | 4.68 |
ENSDART00000138849
|
zgc:56699
|
zgc:56699 |
| chr20_-_26531850 | 3.94 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr20_-_25626693 | 3.50 |
ENSDART00000132247
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr16_-_42186093 | 3.47 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr9_-_11587070 | 3.45 |
ENSDART00000030995
|
umps
|
uridine monophosphate synthetase |
| chr5_+_68807170 | 3.41 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr9_-_12652984 | 3.39 |
ENSDART00000052256
|
sumo3b
|
small ubiquitin-like modifier 3b |
| chr22_+_24220937 | 3.34 |
ENSDART00000165380
|
uchl5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr9_+_38158570 | 3.31 |
ENSDART00000059549
ENSDART00000133060 |
nifk
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr13_+_15701849 | 3.24 |
ENSDART00000003517
|
trmt61a
|
tRNA methyltransferase 61A |
| chr1_-_19648227 | 3.23 |
ENSDART00000054574
|
polr1e
|
polymerase (RNA) I polypeptide E |
| chr8_+_23355484 | 3.22 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr10_+_35002786 | 3.21 |
ENSDART00000099552
|
exosc8
|
exosome component 8 |
| chr7_+_49715750 | 3.01 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr1_+_5485799 | 2.97 |
ENSDART00000022307
|
atic
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr20_+_25626479 | 2.86 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr20_-_25631256 | 2.76 |
ENSDART00000048164
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr22_+_5120033 | 2.73 |
ENSDART00000169200
|
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr13_-_12021566 | 2.72 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr3_+_24595922 | 2.68 |
ENSDART00000169405
|
si:dkey-68o6.5
|
si:dkey-68o6.5 |
| chr8_+_26059677 | 2.67 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr23_-_38160024 | 2.60 |
ENSDART00000087112
|
pfdn4
|
prefoldin subunit 4 |
| chr21_-_30408775 | 2.56 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr13_+_13930263 | 2.54 |
ENSDART00000079154
|
rpia
|
ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) |
| chr14_+_16287968 | 2.48 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr17_-_48705993 | 2.48 |
ENSDART00000030934
|
kcnk5a
|
potassium channel, subfamily K, member 5a |
| chr10_+_22034477 | 2.48 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr17_-_19534474 | 2.44 |
ENSDART00000192469
|
cyp26c1
|
cytochrome P450, family 26, subfamily C, polypeptide 1 |
| chr14_+_14836468 | 2.35 |
ENSDART00000166728
|
si:dkey-102m7.3
|
si:dkey-102m7.3 |
| chr21_-_14832369 | 2.34 |
ENSDART00000144859
|
pus1
|
pseudouridylate synthase 1 |
| chr7_-_60831082 | 2.33 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr21_-_19919918 | 2.33 |
ENSDART00000137307
ENSDART00000142523 ENSDART00000065670 |
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr5_-_55914268 | 2.32 |
ENSDART00000014049
|
wdr36
|
WD repeat domain 36 |
| chr24_-_21923930 | 2.29 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr2_+_12255568 | 2.26 |
ENSDART00000184164
ENSDART00000013454 |
prtfdc1
|
phosphoribosyl transferase domain containing 1 |
| chr14_-_41369629 | 2.25 |
ENSDART00000173040
|
cstf2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2 |
| chr5_-_8765428 | 2.24 |
ENSDART00000167793
|
mybbp1a
|
MYB binding protein (P160) 1a |
| chr11_-_26375575 | 2.24 |
ENSDART00000079255
|
utp3
|
UTP3, small subunit (SSU) processome component, homolog (S. cerevisiae) |
| chr22_+_5118361 | 2.22 |
ENSDART00000168371
ENSDART00000170222 ENSDART00000158846 |
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr19_-_24555935 | 2.21 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr5_-_54714525 | 2.21 |
ENSDART00000150138
ENSDART00000150070 |
ccnb1
|
cyclin B1 |
| chr19_-_24555623 | 2.21 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr13_-_45022301 | 2.20 |
ENSDART00000183589
ENSDART00000125633 ENSDART00000074787 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr16_+_23960933 | 2.18 |
ENSDART00000146077
|
apoeb
|
apolipoprotein Eb |
| chr17_+_53428092 | 2.14 |
ENSDART00000192509
|
srsf10b
|
serine/arginine-rich splicing factor 10b |
| chr5_-_54714789 | 2.14 |
ENSDART00000063357
|
ccnb1
|
cyclin B1 |
| chr1_+_9860381 | 2.12 |
ENSDART00000054848
|
pmm2
|
phosphomannomutase 2 |
| chr14_+_22113331 | 2.11 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr16_+_23960744 | 2.09 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr13_-_4664403 | 2.03 |
ENSDART00000023803
ENSDART00000177957 |
c1d
|
C1D nuclear receptor corepressor |
| chr19_+_10831362 | 2.02 |
ENSDART00000053325
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr12_-_8486330 | 2.02 |
ENSDART00000062855
|
egr2b
|
early growth response 2b |
| chr3_+_23691847 | 2.02 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr20_+_25445826 | 2.02 |
ENSDART00000012581
|
pfas
|
phosphoribosylformylglycinamidine synthase |
| chr22_-_26834043 | 2.02 |
ENSDART00000087202
|
si:dkey-44g23.5
|
si:dkey-44g23.5 |
| chr5_-_10082244 | 2.01 |
ENSDART00000036421
|
chek2
|
checkpoint kinase 2 |
| chr19_+_20778011 | 2.01 |
ENSDART00000024208
|
nutf2l
|
nuclear transport factor 2, like |
| chr2_+_53720028 | 2.01 |
ENSDART00000170799
|
ctnnbl1
|
catenin, beta like 1 |
| chr13_-_4992395 | 1.99 |
ENSDART00000102651
|
nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr6_-_13114821 | 1.97 |
ENSDART00000164579
ENSDART00000165375 |
zgc:194469
|
zgc:194469 |
| chr20_+_46311707 | 1.95 |
ENSDART00000184743
|
flvcr2b
|
feline leukemia virus subgroup C cellular receptor family, member 2b |
| chr13_+_30912385 | 1.95 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
| chr21_-_22122312 | 1.95 |
ENSDART00000101726
|
slc35f2
|
solute carrier family 35, member F2 |
| chr22_+_9922301 | 1.94 |
ENSDART00000105924
|
blf
|
bloody fingers |
| chr21_+_10756154 | 1.94 |
ENSDART00000074833
|
rx3
|
retinal homeobox gene 3 |
| chr23_-_16485190 | 1.93 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr3_-_32362872 | 1.90 |
ENSDART00000035545
ENSDART00000012630 |
prmt1
|
protein arginine methyltransferase 1 |
| chr19_-_30800004 | 1.90 |
ENSDART00000128560
ENSDART00000045504 ENSDART00000125893 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr20_+_25625872 | 1.87 |
ENSDART00000078385
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr23_-_43718067 | 1.85 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr7_-_30779575 | 1.84 |
ENSDART00000004782
|
mphosph10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr4_-_2945306 | 1.83 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr20_-_22484621 | 1.83 |
ENSDART00000063601
|
gsx2
|
GS homeobox 2 |
| chr13_+_30912117 | 1.81 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr13_+_48359573 | 1.81 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr17_+_17764979 | 1.80 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr19_+_20793388 | 1.80 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr2_+_23613040 | 1.77 |
ENSDART00000026694
|
rpp40
|
ribonuclease P/MRP 40 subunit |
| chr10_+_16036573 | 1.77 |
ENSDART00000188757
|
lmnb1
|
lamin B1 |
| chr8_+_29593986 | 1.76 |
ENSDART00000077642
|
atoh1a
|
atonal bHLH transcription factor 1a |
| chr16_+_23961276 | 1.76 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr12_+_31608905 | 1.76 |
ENSDART00000152874
ENSDART00000152996 |
CR352342.1
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr18_-_38245062 | 1.76 |
ENSDART00000189092
|
nat10
|
N-acetyltransferase 10 |
| chr14_-_22113600 | 1.75 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr4_+_17642731 | 1.75 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr6_+_54078703 | 1.75 |
ENSDART00000110845
|
fhit
|
fragile histidine triad gene |
| chr16_+_13860299 | 1.73 |
ENSDART00000121998
|
grwd1
|
glutamate-rich WD repeat containing 1 |
| chr23_+_21459263 | 1.73 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr23_+_36122058 | 1.70 |
ENSDART00000184448
|
hoxc3a
|
homeobox C3a |
| chr11_+_3959495 | 1.70 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr13_+_25200105 | 1.70 |
ENSDART00000039640
|
ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr5_-_1962500 | 1.69 |
ENSDART00000150163
|
rplp0
|
ribosomal protein, large, P0 |
| chr18_-_38244871 | 1.69 |
ENSDART00000076399
|
nat10
|
N-acetyltransferase 10 |
| chr25_-_8625601 | 1.69 |
ENSDART00000155280
|
GDPGP1
|
zgc:153343 |
| chr13_+_48358467 | 1.68 |
ENSDART00000171080
ENSDART00000162531 |
msh6
|
mutS homolog 6 (E. coli) |
| chr16_-_33650578 | 1.67 |
ENSDART00000058460
|
utp11l
|
UTP11-like, U3 small nucleolar ribonucleoprotein (yeast) |
| chr3_+_34120191 | 1.66 |
ENSDART00000020017
ENSDART00000151700 |
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr2_+_20967673 | 1.66 |
ENSDART00000057174
|
arpc5a
|
actin related protein 2/3 complex, subunit 5A |
| chr6_+_3717613 | 1.65 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr9_+_32301456 | 1.64 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr6_+_21001264 | 1.64 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr12_+_18445604 | 1.64 |
ENSDART00000078860
|
noxo1b
|
NADPH oxidase organizer 1b |
| chr7_+_22792132 | 1.63 |
ENSDART00000135207
ENSDART00000146801 |
rbm4.3
|
RNA binding motif protein 4.3 |
| chr2_-_6065416 | 1.63 |
ENSDART00000037698
|
uck2b
|
uridine-cytidine kinase 2b |
| chr16_-_21489514 | 1.63 |
ENSDART00000149525
ENSDART00000148517 ENSDART00000146914 ENSDART00000186493 ENSDART00000193081 ENSDART00000186017 |
mpp6a
|
membrane protein, palmitoylated 6a (MAGUK p55 subfamily member 6) |
| chr19_-_2318391 | 1.62 |
ENSDART00000012791
|
sp8a
|
sp8 transcription factor a |
| chr20_-_38827623 | 1.62 |
ENSDART00000153310
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr6_-_13114406 | 1.62 |
ENSDART00000188015
|
zgc:194469
|
zgc:194469 |
| chr14_+_22076596 | 1.61 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr4_+_61171310 | 1.60 |
ENSDART00000141738
|
si:dkey-9p20.18
|
si:dkey-9p20.18 |
| chr23_-_37085547 | 1.60 |
ENSDART00000144333
|
ints11
|
integrator complex subunit 11 |
| chr1_-_35916247 | 1.60 |
ENSDART00000181541
|
smad1
|
SMAD family member 1 |
| chr13_-_35908275 | 1.60 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr5_+_3927989 | 1.60 |
ENSDART00000030125
|
znhit3
|
zinc finger, HIT-type containing 3 |
| chr25_-_35956344 | 1.59 |
ENSDART00000066987
|
mphosph6
|
M-phase phosphoprotein 6 |
| chr10_+_21434649 | 1.59 |
ENSDART00000193938
ENSDART00000064558 |
etf1b
|
eukaryotic translation termination factor 1b |
| chr23_+_38159715 | 1.59 |
ENSDART00000137969
|
zgc:112994
|
zgc:112994 |
| chr13_-_31370184 | 1.59 |
ENSDART00000034829
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr13_-_25408387 | 1.59 |
ENSDART00000002741
|
itprip
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr20_+_48116476 | 1.58 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr23_+_2906031 | 1.57 |
ENSDART00000109304
|
c23h20orf24
|
c23h20orf24 homolog (H. sapiens) |
| chr3_+_19665319 | 1.57 |
ENSDART00000007857
ENSDART00000193509 |
mettl2a
|
methyltransferase like 2A |
| chr17_-_49407091 | 1.55 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr12_-_34827477 | 1.55 |
ENSDART00000153026
|
NDUFAF8
|
si:dkey-21c1.6 |
| chr18_-_11184584 | 1.55 |
ENSDART00000040500
|
tspan9a
|
tetraspanin 9a |
| chr2_-_32512648 | 1.53 |
ENSDART00000170674
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr24_+_36317544 | 1.53 |
ENSDART00000048640
ENSDART00000156096 |
pus3
|
pseudouridylate synthase 3 |
| chr25_+_19095231 | 1.52 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr7_+_22801465 | 1.52 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr11_+_25485774 | 1.52 |
ENSDART00000026249
|
gnl3l
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr2_-_28102264 | 1.51 |
ENSDART00000013638
|
cdh6
|
cadherin 6 |
| chr13_+_31390313 | 1.51 |
ENSDART00000111477
ENSDART00000137291 |
METTL18
|
si:dkey-15f17.8 |
| chr5_+_25760112 | 1.51 |
ENSDART00000088011
ENSDART00000182046 |
tmem2
|
transmembrane protein 2 |
| chr14_-_25985698 | 1.50 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr2_-_10386738 | 1.50 |
ENSDART00000016369
|
wls
|
wntless Wnt ligand secretion mediator |
| chr9_-_11676491 | 1.50 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr18_-_5850683 | 1.49 |
ENSDART00000082087
|
nip7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr12_-_31724198 | 1.49 |
ENSDART00000153056
ENSDART00000165299 ENSDART00000137464 ENSDART00000080173 |
srsf2a
|
serine/arginine-rich splicing factor 2a |
| chr23_+_31245395 | 1.49 |
ENSDART00000053588
|
irak1bp1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr7_-_39751540 | 1.49 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr22_-_24984733 | 1.49 |
ENSDART00000142147
ENSDART00000187284 |
dnal4b
|
dynein, axonemal, light chain 4b |
| chr24_+_32668675 | 1.48 |
ENSDART00000156638
ENSDART00000155973 |
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr23_+_36653376 | 1.48 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr9_+_907459 | 1.48 |
ENSDART00000034850
ENSDART00000144114 |
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr13_-_35907768 | 1.47 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr25_+_245438 | 1.47 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr10_-_17232372 | 1.46 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr23_+_31912882 | 1.46 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr20_+_38671894 | 1.45 |
ENSDART00000146544
|
mpv17
|
MpV17 mitochondrial inner membrane protein |
| chr19_+_2685779 | 1.45 |
ENSDART00000160533
ENSDART00000097531 |
tomm7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr23_-_37085959 | 1.44 |
ENSDART00000139662
ENSDART00000031016 |
ints11
|
integrator complex subunit 11 |
| chr8_+_2487883 | 1.43 |
ENSDART00000101841
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr14_+_28432737 | 1.43 |
ENSDART00000026846
|
pin4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr1_-_18592068 | 1.41 |
ENSDART00000082063
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr1_-_55750208 | 1.40 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr6_-_52348562 | 1.40 |
ENSDART00000142565
ENSDART00000145369 ENSDART00000016890 |
eif6
|
eukaryotic translation initiation factor 6 |
| chr8_+_25295160 | 1.40 |
ENSDART00000049793
|
gstm.1
|
glutathione S-transferase mu, tandem duplicate 1 |
| chr13_-_25199260 | 1.39 |
ENSDART00000057605
|
adka
|
adenosine kinase a |
| chr16_+_40954481 | 1.39 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr12_-_17602958 | 1.38 |
ENSDART00000134690
ENSDART00000028090 |
eif2ak1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr12_+_27117609 | 1.38 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr2_-_5199431 | 1.38 |
ENSDART00000063384
|
phb2a
|
prohibitin 2a |
| chr23_+_37086159 | 1.38 |
ENSDART00000074407
|
cptp
|
ceramide-1-phosphate transfer protein |
| chr2_+_18988407 | 1.37 |
ENSDART00000170216
|
glula
|
glutamate-ammonia ligase (glutamine synthase) a |
| chr12_+_33975065 | 1.36 |
ENSDART00000036649
|
sfxn2
|
sideroflexin 2 |
| chr8_+_26432677 | 1.36 |
ENSDART00000078369
ENSDART00000131925 |
zgc:136971
|
zgc:136971 |
| chr2_+_49860722 | 1.35 |
ENSDART00000144060
|
rpl37
|
ribosomal protein L37 |
| chr11_+_17984354 | 1.35 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr1_-_40015782 | 1.35 |
ENSDART00000157425
ENSDART00000159238 |
cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr4_+_62341346 | 1.35 |
ENSDART00000160601
|
znf1079
|
zinc finger protein 1079 |
| chr19_+_19750101 | 1.33 |
ENSDART00000168041
ENSDART00000170697 |
hoxa9a
CR382300.2
|
homeobox A9a |
| chr13_+_15702142 | 1.33 |
ENSDART00000135960
|
trmt61a
|
tRNA methyltransferase 61A |
| chr21_+_6114709 | 1.33 |
ENSDART00000065858
|
fpgs
|
folylpolyglutamate synthase |
| chr11_-_30636163 | 1.33 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr11_-_6206520 | 1.32 |
ENSDART00000150199
ENSDART00000148246 ENSDART00000019440 |
pole4
|
polymerase (DNA-directed), epsilon 4, accessory subunit |
| chr3_+_24275766 | 1.32 |
ENSDART00000055607
|
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr7_-_59564011 | 1.31 |
ENSDART00000186053
|
zgc:112271
|
zgc:112271 |
| chr7_+_59169081 | 1.31 |
ENSDART00000167980
|
ostc
|
oligosaccharyltransferase complex subunit |
| chr11_+_6010177 | 1.31 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr18_+_5490668 | 1.31 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr25_-_35296165 | 1.31 |
ENSDART00000018107
|
fancf
|
Fanconi anemia, complementation group F |
| chr9_-_1970071 | 1.30 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr16_+_28728347 | 1.30 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr13_+_25199849 | 1.30 |
ENSDART00000139209
ENSDART00000130876 |
ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr20_+_43648369 | 1.29 |
ENSDART00000187930
ENSDART00000017269 |
parp1
|
poly (ADP-ribose) polymerase 1 |
| chr7_+_41322407 | 1.28 |
ENSDART00000114076
ENSDART00000139093 |
dph2
|
DPH2 homolog (S. cerevisiae) |
| chr5_+_61475451 | 1.28 |
ENSDART00000163444
|
lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr21_-_26495700 | 1.28 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr4_+_18963822 | 1.27 |
ENSDART00000066975
ENSDART00000066973 |
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr1_+_56180416 | 1.26 |
ENSDART00000089358
|
crb3b
|
crumbs homolog 3b |
| chr8_+_7316568 | 1.26 |
ENSDART00000140874
|
selenoh
|
selenoprotein H |
| chr24_-_6678640 | 1.25 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr25_+_3231812 | 1.25 |
ENSDART00000163647
|
metap2b
|
methionyl aminopeptidase 2b |
| chr21_-_2958422 | 1.25 |
ENSDART00000174091
|
zgc:194215
|
zgc:194215 |
| chr23_-_31932076 | 1.25 |
ENSDART00000138617
|
si:dkey-126g1.7
|
si:dkey-126g1.7 |
| chr6_-_18531349 | 1.24 |
ENSDART00000160693
ENSDART00000169780 |
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr19_+_19786117 | 1.24 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr2_+_25839193 | 1.23 |
ENSDART00000078634
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mycn
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 28.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 2.0 | 6.0 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 1.5 | 4.4 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 1.1 | 3.2 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 1.0 | 5.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 1.0 | 3.9 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.9 | 2.6 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.8 | 2.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.8 | 3.0 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.7 | 2.1 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.6 | 3.2 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.6 | 1.9 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.6 | 2.5 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.6 | 1.8 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.5 | 2.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.5 | 2.5 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.5 | 4.6 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.5 | 1.5 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.5 | 3.5 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.5 | 2.0 | GO:0097037 | heme export(GO:0097037) |
| 0.5 | 2.9 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.5 | 4.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.5 | 1.4 | GO:0046048 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.5 | 1.4 | GO:0042256 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.4 | 1.8 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.4 | 1.7 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.4 | 2.5 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.4 | 2.4 | GO:0003151 | outflow tract morphogenesis(GO:0003151) atrial cardiac muscle cell development(GO:0055014) |
| 0.4 | 3.6 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.4 | 1.2 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.4 | 1.1 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.4 | 5.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.4 | 2.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.4 | 3.6 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.4 | 1.8 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.4 | 1.8 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.4 | 2.1 | GO:0006013 | mannose metabolic process(GO:0006013) GDP-mannose biosynthetic process(GO:0009298) |
| 0.3 | 1.4 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.3 | 6.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.3 | 1.0 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.3 | 3.0 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.3 | 5.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.3 | 1.3 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.3 | 1.6 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 1.3 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.3 | 1.8 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.3 | 1.2 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.3 | 1.2 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.3 | 1.7 | GO:0015961 | diadenosine polyphosphate metabolic process(GO:0015959) diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.3 | 2.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.3 | 1.7 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.3 | 0.5 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.3 | 1.3 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.3 | 1.5 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.3 | 1.0 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 | 2.0 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.2 | 0.7 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.2 | 6.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 1.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 0.7 | GO:0021985 | neurohypophysis development(GO:0021985) |
| 0.2 | 8.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 1.9 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.2 | 0.7 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.2 | 0.7 | GO:0060760 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.2 | 1.1 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.2 | 0.6 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.2 | 6.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.2 | 1.8 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 1.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 1.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 1.1 | GO:1902023 | ornithine transport(GO:0015822) L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-ornithine transmembrane transport(GO:1903352) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.2 | 0.8 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 1.1 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.2 | 0.9 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 1.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 1.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 3.4 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.2 | 1.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 1.2 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.2 | 1.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 1.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 | 0.8 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.2 | 1.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 1.6 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.2 | 0.8 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.2 | 2.0 | GO:0048941 | myelination of lateral line nerve axons(GO:0048897) posterior lateral line nerve glial cell differentiation(GO:0048931) myelination of posterior lateral line nerve axons(GO:0048932) lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048938) posterior lateral line nerve glial cell development(GO:0048941) posterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048942) |
| 0.2 | 0.5 | GO:0006585 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.1 | 1.0 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 1.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.6 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.4 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.4 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.1 | 3.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 2.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.5 | GO:0021987 | pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.1 | 3.4 | GO:0001757 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.1 | 1.6 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.7 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 9.9 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.1 | 0.3 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.1 | 0.6 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.8 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 2.4 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 3.2 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 1.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.1 | 1.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 2.5 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 0.4 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 2.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.6 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 0.5 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 2.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 1.0 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 0.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.9 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.1 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 2.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.6 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 2.0 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.1 | 2.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 3.4 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.1 | 0.4 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 1.6 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 3.3 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 1.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 1.1 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 1.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 1.9 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 0.6 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.1 | 1.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.9 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.6 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 1.6 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 1.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.9 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.6 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 1.0 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 1.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 1.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 4.7 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.3 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 1.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.6 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.5 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 1.5 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 1.6 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 2.2 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.3 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 1.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.6 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 1.4 | GO:1990542 | mitochondrial transmembrane transport(GO:1990542) |
| 0.0 | 3.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 1.6 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 1.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.2 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.7 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 1.0 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.5 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 1.0 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.9 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 6.8 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.2 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 2.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.5 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.3 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.5 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.2 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.4 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.4 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.3 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 2.3 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 1.2 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.0 | 2.0 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 2.2 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 1.1 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.4 | 4.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 1.2 | 3.5 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.9 | 4.7 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.6 | 2.6 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.6 | 1.8 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.6 | 3.4 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.6 | 3.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.5 | 3.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.4 | 1.8 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.4 | 6.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.4 | 1.2 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.4 | 4.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.4 | 1.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.3 | 1.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.3 | 1.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.3 | 1.6 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.3 | 3.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.3 | 0.9 | GO:0031362 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 1.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 3.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.3 | 2.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.3 | 1.8 | GO:0030681 | ribonuclease MRP complex(GO:0000172) multimeric ribonuclease P complex(GO:0030681) |
| 0.3 | 1.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 1.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.2 | 1.2 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.2 | 2.0 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 3.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.2 | 0.8 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 4.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.2 | 4.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 3.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 1.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.0 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 1.1 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 2.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.5 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 1.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.8 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 1.8 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.8 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 2.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 3.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 25.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.1 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 2.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.6 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 1.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.4 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 1.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.6 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.1 | 3.3 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.2 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 5.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 1.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 5.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 30.1 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 2.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 3.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 8.4 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.0 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 18.9 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 2.0 | 6.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 1.2 | 3.5 | GO:0032405 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.9 | 4.7 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.9 | 6.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.9 | 2.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.8 | 5.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.8 | 2.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.8 | 2.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.7 | 3.9 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.6 | 3.0 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.5 | 1.6 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.5 | 1.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.5 | 1.8 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.4 | 1.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 1.6 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.4 | 3.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 6.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.3 | 1.0 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.3 | 1.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 0.9 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.3 | 1.5 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.3 | 1.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 1.4 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.3 | 2.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.3 | 1.4 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.3 | 0.8 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.3 | 0.8 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 2.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 3.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.2 | 6.7 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.2 | 3.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 2.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 3.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 1.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 1.4 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 1.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 3.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 1.9 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.2 | 2.9 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.2 | 3.1 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.2 | 0.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 0.6 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 1.4 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 0.8 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 3.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.2 | 1.6 | GO:0016176 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 2.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 2.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.2 | 1.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 1.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 0.5 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 2.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 1.5 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 1.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.6 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.5 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 2.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.7 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.9 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 0.7 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 1.7 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.3 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.4 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.1 | 4.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.4 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.1 | 2.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 3.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.4 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 1.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 0.5 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 2.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 1.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 1.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 2.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 3.4 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 2.2 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 2.1 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 2.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.1 | 4.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 6.0 | GO:0051540 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.1 | 1.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 2.6 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.1 | 1.0 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.9 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 1.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 1.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 1.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.9 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 1.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 1.9 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 2.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 1.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 2.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 1.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.3 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.6 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.6 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 11.4 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.0 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 3.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 14.7 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.9 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 1.0 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.8 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 6.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 3.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 2.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 31.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.5 | 6.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 5.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 2.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 1.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 3.3 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.2 | 3.0 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 2.0 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 2.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.1 | 1.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 3.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 2.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 3.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 5.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.7 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 0.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.2 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.1 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.9 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 2.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 2.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 2.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 3.3 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 1.2 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.6 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME DNA STRAND ELONGATION | Genes involved in DNA strand elongation |
| 0.0 | 1.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |