Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
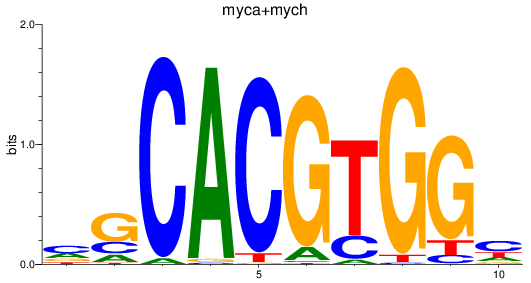
Results for myca+mych
Z-value: 0.68
Transcription factors associated with myca+mych
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myca
|
ENSDARG00000045695 | MYC proto-oncogene, bHLH transcription factor a |
|
mych
|
ENSDARG00000077473 | myelocytomatosis oncogene homolog |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myca | dr11_v1_chr24_+_10414028_10414028 | 0.86 | 2.8e-06 | Click! |
| mych | dr11_v1_chr6_+_50451337_50451337 | -0.03 | 8.9e-01 | Click! |
Activity profile of myca+mych motif
Sorted Z-values of myca+mych motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_26531850 | 2.69 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr22_+_5118361 | 2.01 |
ENSDART00000168371
ENSDART00000170222 ENSDART00000158846 |
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr20_-_25626198 | 1.87 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr13_-_35892051 | 1.79 |
ENSDART00000145884
|
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr23_+_21459263 | 1.77 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr5_+_68807170 | 1.75 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr13_-_25199260 | 1.54 |
ENSDART00000057605
|
adka
|
adenosine kinase a |
| chr7_+_49715750 | 1.52 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr17_-_19534474 | 1.49 |
ENSDART00000192469
|
cyp26c1
|
cytochrome P450, family 26, subfamily C, polypeptide 1 |
| chr14_-_12822 | 1.48 |
ENSDART00000180650
ENSDART00000188819 |
msx1a
|
muscle segment homeobox 1a |
| chr13_+_48359573 | 1.47 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr13_-_45022301 | 1.44 |
ENSDART00000183589
ENSDART00000125633 ENSDART00000074787 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr20_+_25445826 | 1.42 |
ENSDART00000012581
|
pfas
|
phosphoribosylformylglycinamidine synthase |
| chr20_-_25626428 | 1.37 |
ENSDART00000136475
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr12_+_27117609 | 1.32 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr1_-_19648227 | 1.29 |
ENSDART00000054574
|
polr1e
|
polymerase (RNA) I polypeptide E |
| chr9_-_11587070 | 1.28 |
ENSDART00000030995
|
umps
|
uridine monophosphate synthetase |
| chr7_-_23745984 | 1.24 |
ENSDART00000048050
|
ITGB1BP2
|
zgc:92429 |
| chr8_+_26059677 | 1.22 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr1_-_35916247 | 1.22 |
ENSDART00000181541
|
smad1
|
SMAD family member 1 |
| chr3_+_24595922 | 1.20 |
ENSDART00000169405
|
si:dkey-68o6.5
|
si:dkey-68o6.5 |
| chr9_+_38158570 | 1.20 |
ENSDART00000059549
ENSDART00000133060 |
nifk
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr20_-_53996193 | 1.18 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr20_+_25625872 | 1.18 |
ENSDART00000078385
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr18_-_127873 | 1.18 |
ENSDART00000148490
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr5_-_8765428 | 1.16 |
ENSDART00000167793
|
mybbp1a
|
MYB binding protein (P160) 1a |
| chr13_-_12021566 | 1.13 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr10_+_35002786 | 1.13 |
ENSDART00000099552
|
exosc8
|
exosome component 8 |
| chr22_+_5120033 | 1.12 |
ENSDART00000169200
|
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr9_-_11676491 | 1.11 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr13_+_30912117 | 1.11 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr20_-_25626693 | 1.10 |
ENSDART00000132247
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr10_+_22034477 | 1.09 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr14_+_16287968 | 1.08 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr19_-_24555623 | 1.08 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr13_+_30912385 | 1.08 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
| chr9_-_31278048 | 1.06 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr16_-_42186093 | 1.05 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr20_-_25631256 | 1.05 |
ENSDART00000048164
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr3_-_25377163 | 1.04 |
ENSDART00000055490
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr24_+_41690545 | 1.04 |
ENSDART00000160069
|
lama1
|
laminin, alpha 1 |
| chr19_+_20793388 | 1.04 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr6_+_3717613 | 1.03 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr14_+_22076596 | 1.02 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr23_-_21535040 | 1.02 |
ENSDART00000010647
|
rcc2
|
regulator of chromosome condensation 2 |
| chr14_-_41369629 | 1.01 |
ENSDART00000173040
|
cstf2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2 |
| chr4_-_2945306 | 0.99 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr12_+_33975065 | 0.99 |
ENSDART00000036649
|
sfxn2
|
sideroflexin 2 |
| chr18_-_5850683 | 0.99 |
ENSDART00000082087
|
nip7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr20_-_22484621 | 0.99 |
ENSDART00000063601
|
gsx2
|
GS homeobox 2 |
| chr8_+_23355484 | 0.98 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr1_-_30762264 | 0.97 |
ENSDART00000085454
|
dis3
|
DIS3 exosome endoribonuclease and 3'-5' exoribonuclease |
| chr21_-_30408775 | 0.96 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr1_+_5485799 | 0.96 |
ENSDART00000022307
|
atic
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr23_-_38160024 | 0.96 |
ENSDART00000087112
|
pfdn4
|
prefoldin subunit 4 |
| chr19_-_18136410 | 0.96 |
ENSDART00000012352
|
cbx3a
|
chromobox homolog 3a (HP1 gamma homolog, Drosophila) |
| chr23_+_35714574 | 0.96 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr7_+_54605640 | 0.95 |
ENSDART00000168474
|
fgf3
|
fibroblast growth factor 3 |
| chr17_-_42213285 | 0.95 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr4_+_18963822 | 0.95 |
ENSDART00000066975
ENSDART00000066973 |
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr5_-_55914268 | 0.93 |
ENSDART00000014049
|
wdr36
|
WD repeat domain 36 |
| chr25_+_19095231 | 0.93 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr9_+_27720428 | 0.92 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr3_-_12930217 | 0.91 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr1_-_156375 | 0.91 |
ENSDART00000160221
|
pcid2
|
PCI domain containing 2 |
| chr10_+_39283985 | 0.91 |
ENSDART00000016464
|
dcps
|
decapping enzyme, scavenger |
| chr23_-_21534455 | 0.91 |
ENSDART00000139092
|
rcc2
|
regulator of chromosome condensation 2 |
| chr12_+_18445604 | 0.90 |
ENSDART00000078860
|
noxo1b
|
NADPH oxidase organizer 1b |
| chr14_-_25935167 | 0.90 |
ENSDART00000139855
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr16_+_23961276 | 0.90 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr14_+_16151636 | 0.89 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr3_-_37148594 | 0.88 |
ENSDART00000140855
|
mlx
|
MLX, MAX dimerization protein |
| chr23_+_36122058 | 0.88 |
ENSDART00000184448
|
hoxc3a
|
homeobox C3a |
| chr8_+_26432677 | 0.88 |
ENSDART00000078369
ENSDART00000131925 |
zgc:136971
|
zgc:136971 |
| chr8_+_15277874 | 0.87 |
ENSDART00000146965
|
dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr9_+_36967294 | 0.86 |
ENSDART00000059753
|
inhbb
|
inhibin, beta B |
| chr6_-_27108844 | 0.86 |
ENSDART00000073883
|
dtymk
|
deoxythymidylate kinase (thymidylate kinase) |
| chr19_-_24555935 | 0.86 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr16_-_10223741 | 0.86 |
ENSDART00000188099
|
si:rp71-15i12.1
|
si:rp71-15i12.1 |
| chr13_+_25200105 | 0.86 |
ENSDART00000039640
|
ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr5_+_3927989 | 0.86 |
ENSDART00000030125
|
znhit3
|
zinc finger, HIT-type containing 3 |
| chr24_+_36317544 | 0.85 |
ENSDART00000048640
ENSDART00000156096 |
pus3
|
pseudouridylate synthase 3 |
| chr20_+_25626479 | 0.83 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr25_+_3231812 | 0.82 |
ENSDART00000163647
|
metap2b
|
methionyl aminopeptidase 2b |
| chr23_+_30730121 | 0.82 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr21_-_26495700 | 0.82 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr17_+_23968214 | 0.81 |
ENSDART00000183053
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr7_+_22801465 | 0.81 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr1_+_26110985 | 0.81 |
ENSDART00000054208
|
mtap
|
methylthioadenosine phosphorylase |
| chr5_-_10082244 | 0.81 |
ENSDART00000036421
|
chek2
|
checkpoint kinase 2 |
| chr22_+_15343953 | 0.81 |
ENSDART00000045682
|
rrp36
|
ribosomal RNA processing 36 |
| chr9_-_12652984 | 0.80 |
ENSDART00000052256
|
sumo3b
|
small ubiquitin-like modifier 3b |
| chr13_-_35907768 | 0.80 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr4_+_61171310 | 0.80 |
ENSDART00000141738
|
si:dkey-9p20.18
|
si:dkey-9p20.18 |
| chr14_+_5385855 | 0.79 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr13_-_31370184 | 0.79 |
ENSDART00000034829
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr20_+_1349043 | 0.79 |
ENSDART00000186375
|
tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr17_-_49407091 | 0.77 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr9_-_30259295 | 0.77 |
ENSDART00000139106
|
si:dkey-100n23.5
|
si:dkey-100n23.5 |
| chr12_-_9700605 | 0.76 |
ENSDART00000161063
|
heatr1
|
HEAT repeat containing 1 |
| chr21_+_6114709 | 0.75 |
ENSDART00000065858
|
fpgs
|
folylpolyglutamate synthase |
| chr9_-_27396404 | 0.75 |
ENSDART00000136412
ENSDART00000101401 |
tex30
|
testis expressed 30 |
| chr25_-_8513255 | 0.75 |
ENSDART00000150129
|
polg
|
polymerase (DNA directed), gamma |
| chr25_+_21098990 | 0.75 |
ENSDART00000017488
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr15_-_44052927 | 0.73 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr9_-_27719998 | 0.73 |
ENSDART00000161068
ENSDART00000148195 ENSDART00000138386 |
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr4_-_26032741 | 0.72 |
ENSDART00000188058
|
usp44
|
ubiquitin specific peptidase 44 |
| chr24_-_16905018 | 0.72 |
ENSDART00000066759
|
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr8_-_39977026 | 0.71 |
ENSDART00000141707
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr21_-_22122312 | 0.71 |
ENSDART00000101726
|
slc35f2
|
solute carrier family 35, member F2 |
| chr16_-_8520474 | 0.71 |
ENSDART00000137365
|
grb10b
|
growth factor receptor-bound protein 10b |
| chr11_+_6009984 | 0.70 |
ENSDART00000185680
|
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr14_+_94603 | 0.70 |
ENSDART00000162480
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr17_-_42213822 | 0.69 |
ENSDART00000187904
ENSDART00000180029 |
nkx2.2a
|
NK2 homeobox 2a |
| chr25_+_21098675 | 0.66 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr4_-_64709908 | 0.66 |
ENSDART00000161032
|
si:dkey-9i5.2
|
si:dkey-9i5.2 |
| chr4_-_38033800 | 0.66 |
ENSDART00000159662
|
si:dkeyp-82b4.4
|
si:dkeyp-82b4.4 |
| chr11_+_3959495 | 0.65 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr3_+_18807524 | 0.65 |
ENSDART00000055757
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr6_-_8264751 | 0.64 |
ENSDART00000091628
|
ccdc151
|
coiled-coil domain containing 151 |
| chr14_+_34514336 | 0.64 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr11_+_6010177 | 0.63 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr19_-_46957968 | 0.63 |
ENSDART00000043713
|
angpt1
|
angiopoietin 1 |
| chr24_+_16905188 | 0.63 |
ENSDART00000066760
|
cct5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr25_+_245438 | 0.63 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr18_-_11184584 | 0.62 |
ENSDART00000040500
|
tspan9a
|
tetraspanin 9a |
| chr21_-_30082414 | 0.62 |
ENSDART00000157307
ENSDART00000155188 |
ccnjl
|
cyclin J-like |
| chr16_-_31790285 | 0.62 |
ENSDART00000184655
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr11_+_25485774 | 0.61 |
ENSDART00000026249
|
gnl3l
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr16_+_16968682 | 0.61 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr2_+_25839193 | 0.61 |
ENSDART00000078634
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr1_-_54972170 | 0.60 |
ENSDART00000150548
ENSDART00000038330 |
khsrp
|
KH-type splicing regulatory protein |
| chr23_+_38159715 | 0.60 |
ENSDART00000137969
|
zgc:112994
|
zgc:112994 |
| chr16_+_25761101 | 0.60 |
ENSDART00000110619
|
cblc
|
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr8_-_30979494 | 0.60 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr9_-_27720612 | 0.60 |
ENSDART00000000566
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr16_-_38629208 | 0.59 |
ENSDART00000126705
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr23_+_32499916 | 0.59 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr13_-_25408387 | 0.58 |
ENSDART00000002741
|
itprip
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr24_+_24014880 | 0.58 |
ENSDART00000041335
|
chodl
|
chondrolectin |
| chr9_+_41821613 | 0.57 |
ENSDART00000097295
|
col18a1a
|
collagen type XVIII alpha 1 chain a |
| chr13_-_4992395 | 0.57 |
ENSDART00000102651
|
nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr21_+_25068215 | 0.56 |
ENSDART00000167523
ENSDART00000189259 |
dixdc1b
|
DIX domain containing 1b |
| chr2_-_11027258 | 0.56 |
ENSDART00000081072
ENSDART00000193824 ENSDART00000187036 ENSDART00000097741 |
ssbp3a
|
single stranded DNA binding protein 3a |
| chr19_-_34117056 | 0.56 |
ENSDART00000158677
|
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr19_+_20201254 | 0.56 |
ENSDART00000010140
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr1_-_25438934 | 0.55 |
ENSDART00000111686
|
fhdc1
|
FH2 domain containing 1 |
| chr14_-_25985698 | 0.55 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr3_+_28939759 | 0.54 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr4_-_25271455 | 0.54 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr16_+_16969060 | 0.54 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr6_-_32726848 | 0.53 |
ENSDART00000155294
|
zc3h3
|
zinc finger CCCH-type containing 3 |
| chr7_-_30779575 | 0.53 |
ENSDART00000004782
|
mphosph10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr20_-_48470599 | 0.53 |
ENSDART00000166857
|
CABZ01059120.1
|
|
| chr13_-_9295890 | 0.53 |
ENSDART00000110680
|
HTRA2 (1 of many)
|
si:dkey-33c12.12 |
| chr6_-_18531349 | 0.52 |
ENSDART00000160693
ENSDART00000169780 |
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr25_-_14424406 | 0.52 |
ENSDART00000073609
|
prmt7
|
protein arginine methyltransferase 7 |
| chr16_+_33593116 | 0.51 |
ENSDART00000013148
|
pou3f1
|
POU class 3 homeobox 1 |
| chr18_+_14645568 | 0.51 |
ENSDART00000138995
ENSDART00000147351 |
vps9d1
|
VPS9 domain containing 1 |
| chr14_+_28432737 | 0.50 |
ENSDART00000026846
|
pin4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr19_+_20201593 | 0.50 |
ENSDART00000163026
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr17_+_24843401 | 0.50 |
ENSDART00000110179
|
cx34.4
|
connexin 34.4 |
| chr19_-_15855427 | 0.50 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr21_-_41147818 | 0.50 |
ENSDART00000167339
ENSDART00000192730 |
msx2b
|
muscle segment homeobox 2b |
| chr5_-_3927692 | 0.49 |
ENSDART00000146840
ENSDART00000058346 |
c1qbp
|
complement component 1, q subcomponent binding protein |
| chr1_-_55750208 | 0.47 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr23_-_21534738 | 0.46 |
ENSDART00000134587
|
rcc2
|
regulator of chromosome condensation 2 |
| chr15_-_1484795 | 0.46 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr7_-_55539738 | 0.46 |
ENSDART00000168721
ENSDART00000013796 ENSDART00000148514 |
aprt
|
adenine phosphoribosyltransferase |
| chr4_+_53976731 | 0.45 |
ENSDART00000165813
|
si:ch211-249c2.1
|
si:ch211-249c2.1 |
| chr6_+_54078703 | 0.45 |
ENSDART00000110845
|
fhit
|
fragile histidine triad gene |
| chr7_+_30779761 | 0.44 |
ENSDART00000066806
ENSDART00000173671 |
mcee
|
methylmalonyl CoA epimerase |
| chr17_-_7028418 | 0.44 |
ENSDART00000188305
ENSDART00000187895 |
sash1b
|
SAM and SH3 domain containing 1b |
| chr6_-_58975010 | 0.44 |
ENSDART00000144911
ENSDART00000144514 |
mars
|
methionyl-tRNA synthetase |
| chr2_-_59285085 | 0.44 |
ENSDART00000131880
|
ftr34
|
finTRIM family, member 34 |
| chr9_-_21460164 | 0.43 |
ENSDART00000133469
|
zmym2
|
zinc finger, MYM-type 2 |
| chr8_+_25295160 | 0.43 |
ENSDART00000049793
|
gstm.1
|
glutathione S-transferase mu, tandem duplicate 1 |
| chr13_+_39277178 | 0.43 |
ENSDART00000113259
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr13_+_25199849 | 0.42 |
ENSDART00000139209
ENSDART00000130876 |
ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr1_-_54971968 | 0.42 |
ENSDART00000140016
|
khsrp
|
KH-type splicing regulatory protein |
| chr18_-_20608025 | 0.42 |
ENSDART00000090156
ENSDART00000151980 |
bcl2l13
|
BCL2 like 13 |
| chr21_-_11199366 | 0.42 |
ENSDART00000167666
|
dnajc21
|
DnaJ (Hsp40) homolog, subfamily C, member 21 |
| chr17_+_51682429 | 0.42 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr21_-_28920245 | 0.41 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr8_-_52143109 | 0.41 |
ENSDART00000142969
|
tcf7l1b
|
transcription factor 7 like 1b |
| chr3_+_54887782 | 0.40 |
ENSDART00000018205
|
aanat2
|
arylalkylamine N-acetyltransferase 2 |
| chr22_-_26834043 | 0.40 |
ENSDART00000087202
|
si:dkey-44g23.5
|
si:dkey-44g23.5 |
| chr18_+_6638726 | 0.40 |
ENSDART00000142755
ENSDART00000167781 |
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr14_+_14836468 | 0.40 |
ENSDART00000166728
|
si:dkey-102m7.3
|
si:dkey-102m7.3 |
| chr24_+_23791758 | 0.40 |
ENSDART00000066655
ENSDART00000146580 |
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr6_-_21189295 | 0.39 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr7_-_58843193 | 0.39 |
ENSDART00000167231
|
mrpl15
|
mitochondrial ribosomal protein L15 |
| chr6_-_39080630 | 0.39 |
ENSDART00000021520
ENSDART00000128308 |
eif4bb
|
eukaryotic translation initiation factor 4Bb |
| chr24_+_32668675 | 0.38 |
ENSDART00000156638
ENSDART00000155973 |
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr6_-_52348562 | 0.38 |
ENSDART00000142565
ENSDART00000145369 ENSDART00000016890 |
eif6
|
eukaryotic translation initiation factor 6 |
| chr3_-_58165254 | 0.38 |
ENSDART00000093031
|
snu13a
|
SNU13 homolog, small nuclear ribonucleoprotein a (U4/U6.U5) |
| chr16_+_32136550 | 0.38 |
ENSDART00000147526
|
sphk2
|
sphingosine kinase 2 |
| chr9_-_785444 | 0.37 |
ENSDART00000012506
|
en1a
|
engrailed homeobox 1a |
| chr14_+_94946 | 0.36 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr7_+_52887701 | 0.36 |
ENSDART00000109973
|
tp53bp1
|
tumor protein p53 binding protein, 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of myca+mych
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 9.8 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.8 | 0.8 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.5 | 1.6 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.5 | 1.5 | GO:0006290 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.5 | 1.4 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.4 | 1.3 | GO:0001113 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.4 | 1.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.4 | 1.1 | GO:0071042 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.3 | 2.0 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.3 | 1.0 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.3 | 1.0 | GO:0021985 | neurohypophysis development(GO:0021985) |
| 0.3 | 0.9 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.3 | 0.9 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.3 | 0.9 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.3 | 1.2 | GO:1904407 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.3 | 0.9 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.3 | 1.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.3 | 1.3 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.3 | 1.3 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.2 | 1.5 | GO:0055014 | outflow tract morphogenesis(GO:0003151) atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 3.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 2.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.2 | 0.9 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 1.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 0.8 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.2 | 0.8 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 0.7 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.2 | 0.5 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.2 | 1.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.2 | 0.7 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.2 | 0.6 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.2 | 0.6 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 0.8 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.2 | 1.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.2 | 0.8 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.2 | 1.1 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.1 | 1.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 1.0 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 0.9 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.4 | GO:1902626 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.8 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.5 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.9 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.7 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.1 | 1.0 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.9 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 2.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.4 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 | 0.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.8 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 1.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 1.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.6 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.8 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 0.4 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.1 | 0.2 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.1 | 1.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.4 | GO:0015961 | diadenosine polyphosphate metabolic process(GO:0015959) diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.1 | 1.8 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.6 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.9 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 2.1 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.8 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.1 | 0.6 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.1 | 1.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.4 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.2 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 0.2 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.7 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.4 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 1.5 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 4.0 | GO:0016072 | rRNA processing(GO:0006364) rRNA metabolic process(GO:0016072) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 1.5 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.0 | 0.9 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 1.0 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.4 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.4 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.2 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.0 | 0.8 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.0 | 0.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 1.7 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 1.8 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.8 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.4 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.6 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.6 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.3 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 1.1 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.1 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.2 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 3.0 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 1.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.8 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.6 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.8 | GO:1990542 | mitochondrial transmembrane transport(GO:1990542) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.4 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 0.3 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.3 | 1.3 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.2 | 1.0 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 1.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 1.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 0.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 2.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 1.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 2.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 0.9 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.2 | 0.5 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 2.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.6 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.1 | 0.8 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 0.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.0 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.5 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.0 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 2.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 1.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 7.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.1 | GO:0070938 | actomyosin contractile ring(GO:0005826) contractile ring(GO:0070938) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.4 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.5 | 3.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.5 | 1.5 | GO:0032143 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.5 | 3.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.4 | 2.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 1.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.3 | 1.5 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.3 | 0.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 0.9 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.3 | 0.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 0.9 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.2 | 1.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.2 | 0.6 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.2 | 1.0 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.2 | 0.5 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 1.0 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 0.5 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.1 | 0.4 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.4 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.1 | 0.9 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 0.5 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 1.0 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.4 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 2.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.7 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 1.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.7 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 1.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 2.1 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.4 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.4 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 3.3 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.3 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 3.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 1.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 1.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.6 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 11.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 3.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 0.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.8 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.8 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 0.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.0 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.0 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 2.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.9 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |