Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
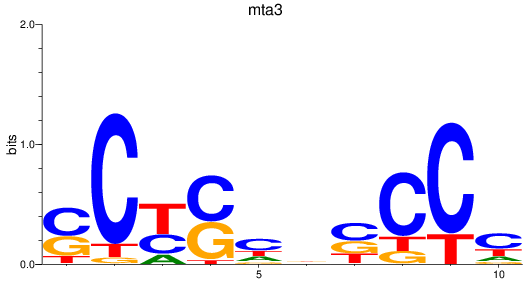
Results for mta3
Z-value: 0.25
Transcription factors associated with mta3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mta3
|
ENSDARG00000054903 | metastasis associated 1 family, member 3 |
|
mta3
|
ENSDARG00000113436 | metastasis associated 1 family, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mta3 | dr11_v1_chr12_+_25223843_25223873 | -0.48 | 3.7e-02 | Click! |
Activity profile of mta3 motif
Sorted Z-values of mta3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_43056153 | 1.01 |
ENSDART00000186377
|
prnpa
|
prion protein a |
| chr21_+_25187210 | 0.84 |
ENSDART00000101147
ENSDART00000167528 |
si:dkey-183i3.5
|
si:dkey-183i3.5 |
| chr2_-_22535 | 0.61 |
ENSDART00000157877
|
CABZ01092282.1
|
|
| chr5_-_20678300 | 0.47 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr13_+_1100197 | 0.43 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr11_-_44543082 | 0.42 |
ENSDART00000099568
|
gpr137bb
|
G protein-coupled receptor 137Bb |
| chr3_+_16566361 | 0.41 |
ENSDART00000132869
ENSDART00000080771 |
slc6a16a
|
solute carrier family 6, member 16a |
| chr2_-_43168292 | 0.41 |
ENSDART00000132588
|
crema
|
cAMP responsive element modulator a |
| chr14_-_572742 | 0.40 |
ENSDART00000168951
|
fgf2
|
fibroblast growth factor 2 |
| chr11_-_32723851 | 0.37 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr14_-_2369849 | 0.37 |
ENSDART00000180422
ENSDART00000189731 ENSDART00000111748 |
pcdhb
|
protocadherin b |
| chr16_-_17207754 | 0.37 |
ENSDART00000063804
|
wu:fj39g12
|
wu:fj39g12 |
| chr7_-_6592142 | 0.34 |
ENSDART00000160137
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr21_+_44581654 | 0.30 |
ENSDART00000187201
ENSDART00000180360 ENSDART00000191543 ENSDART00000180039 ENSDART00000186308 |
TMEM164
|
transmembrane protein 164 |
| chr10_-_9115383 | 0.29 |
ENSDART00000139324
|
si:dkeyp-41f9.3
|
si:dkeyp-41f9.3 |
| chr23_-_24825863 | 0.27 |
ENSDART00000112493
|
syt6a
|
synaptotagmin VIa |
| chr20_-_51727860 | 0.25 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr12_-_14551077 | 0.25 |
ENSDART00000188717
|
FO704673.1
|
|
| chr16_-_280835 | 0.25 |
ENSDART00000190541
|
LO017917.1
|
|
| chr7_+_1579510 | 0.25 |
ENSDART00000190525
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr5_+_45138934 | 0.21 |
ENSDART00000041412
ENSDART00000136002 |
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_-_5228137 | 0.18 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr9_-_1434484 | 0.17 |
ENSDART00000093412
|
osbpl6
|
oxysterol binding protein-like 6 |
| chr5_-_25123807 | 0.16 |
ENSDART00000183171
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr20_-_33675676 | 0.16 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr5_+_45139196 | 0.15 |
ENSDART00000113738
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_-_56095275 | 0.15 |
ENSDART00000154701
|
si:ch211-178n15.1
|
si:ch211-178n15.1 |
| chr2_-_16359042 | 0.14 |
ENSDART00000057216
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr19_+_41769135 | 0.14 |
ENSDART00000087096
|
tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr13_+_31545812 | 0.12 |
ENSDART00000076527
|
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr2_+_13069168 | 0.09 |
ENSDART00000192832
|
prkag2b
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit b |
| chr10_-_43928937 | 0.09 |
ENSDART00000168014
ENSDART00000086220 |
mctp1b
|
multiple C2 domains, transmembrane 1b |
| chr13_+_23093743 | 0.08 |
ENSDART00000148034
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr16_+_11029762 | 0.08 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr25_-_35106751 | 0.05 |
ENSDART00000180682
ENSDART00000073457 |
zgc:194989
|
zgc:194989 |
| chr22_-_7809904 | 0.05 |
ENSDART00000182269
ENSDART00000097201 ENSDART00000138522 |
si:ch73-44m9.3
si:ch73-44m9.1
|
si:ch73-44m9.3 si:ch73-44m9.1 |
| chr16_+_13822137 | 0.02 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr4_-_13255700 | 0.01 |
ENSDART00000162277
ENSDART00000026593 |
grip1
|
glutamate receptor interacting protein 1 |
| chr4_+_77971104 | 0.00 |
ENSDART00000188609
|
zgc:113921
|
zgc:113921 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mta3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 0.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.4 | GO:0030323 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |