Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
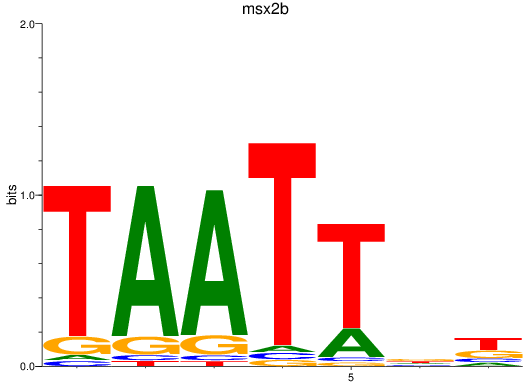
Results for msx2b
Z-value: 0.42
Transcription factors associated with msx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msx2b
|
ENSDARG00000101023 | muscle segment homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msx2b | dr11_v1_chr21_-_41147818_41147818 | -0.49 | 3.4e-02 | Click! |
Activity profile of msx2b motif
Sorted Z-values of msx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_11385155 | 1.67 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr3_+_36366577 | 1.28 |
ENSDART00000151203
|
rgs9a
|
regulator of G protein signaling 9a |
| chr17_-_6730247 | 1.12 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr15_-_34408777 | 1.04 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr10_+_25726694 | 1.02 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr20_+_35484070 | 0.95 |
ENSDART00000026234
ENSDART00000141675 |
mep1a.2
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 2 |
| chr13_+_23988442 | 0.92 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr6_-_46861676 | 0.86 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr23_+_40460333 | 0.76 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr19_-_47527093 | 0.74 |
ENSDART00000171379
ENSDART00000171140 ENSDART00000114886 |
scg5
|
secretogranin V |
| chr22_+_17203752 | 0.69 |
ENSDART00000143376
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr1_+_42225060 | 0.68 |
ENSDART00000138740
ENSDART00000101306 |
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr13_+_8958438 | 0.64 |
ENSDART00000165721
ENSDART00000169398 |
haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr20_-_29474859 | 0.62 |
ENSDART00000152906
ENSDART00000045249 |
scg5
|
secretogranin V |
| chr1_+_11977426 | 0.61 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr8_+_40477264 | 0.60 |
ENSDART00000085559
|
gck
|
glucokinase (hexokinase 4) |
| chr17_-_200316 | 0.60 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr14_-_49063157 | 0.60 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr23_-_24493768 | 0.60 |
ENSDART00000135559
ENSDART00000046933 |
sult1st5
|
sulfotransferase family 1, cytosolic sulfotransferase 5 |
| chr24_-_14712427 | 0.60 |
ENSDART00000176316
|
jph1a
|
junctophilin 1a |
| chr12_+_28574863 | 0.58 |
ENSDART00000153284
|
tbkbp1
|
TBK1 binding protein 1 |
| chr11_+_29770966 | 0.58 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr2_+_53357953 | 0.56 |
ENSDART00000187577
|
celf5b
|
cugbp, Elav-like family member 5b |
| chr13_+_2625150 | 0.56 |
ENSDART00000164177
|
plpp4
|
phospholipid phosphatase 4 |
| chr1_+_34203817 | 0.55 |
ENSDART00000191432
ENSDART00000046094 |
arl6
|
ADP-ribosylation factor-like 6 |
| chr6_-_30210378 | 0.54 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr10_-_2522588 | 0.54 |
ENSDART00000081926
|
CU856539.1
|
|
| chr17_+_31185276 | 0.52 |
ENSDART00000062887
|
disp2
|
dispatched homolog 2 (Drosophila) |
| chr7_+_65309738 | 0.50 |
ENSDART00000169228
|
vat1l
|
vesicle amine transport 1-like |
| chr7_+_69356661 | 0.47 |
ENSDART00000177049
|
CABZ01083862.1
|
|
| chr20_+_4392687 | 0.47 |
ENSDART00000187271
|
im:7142702
|
im:7142702 |
| chr8_-_44004135 | 0.46 |
ENSDART00000136269
|
rimbp2
|
RIMS binding protein 2 |
| chr24_-_24849091 | 0.46 |
ENSDART00000133649
ENSDART00000038290 |
crhb
|
corticotropin releasing hormone b |
| chr17_-_51199558 | 0.46 |
ENSDART00000154532
|
paplna
|
papilin a, proteoglycan-like sulfated glycoprotein |
| chr20_-_53366137 | 0.44 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr21_-_39639954 | 0.44 |
ENSDART00000026766
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr25_+_5604512 | 0.44 |
ENSDART00000042781
|
plxnb2b
|
plexin b2b |
| chr21_+_21195487 | 0.43 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr2_-_23778180 | 0.43 |
ENSDART00000136782
|
si:dkey-24c2.7
|
si:dkey-24c2.7 |
| chr3_+_22935183 | 0.42 |
ENSDART00000157378
|
hdac5
|
histone deacetylase 5 |
| chr21_-_44009169 | 0.42 |
ENSDART00000028960
|
ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| chr15_+_19646902 | 0.42 |
ENSDART00000179822
|
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr13_-_36050303 | 0.40 |
ENSDART00000134955
ENSDART00000139087 |
lgmn
|
legumain |
| chr8_+_22930627 | 0.40 |
ENSDART00000187860
|
sypa
|
synaptophysin a |
| chr23_+_13375359 | 0.39 |
ENSDART00000151947
|
ZNF512B
|
si:dkey-256k13.2 |
| chr3_-_43821191 | 0.39 |
ENSDART00000186816
|
snn
|
stannin |
| chr18_+_10884996 | 0.38 |
ENSDART00000147613
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr16_+_13818500 | 0.38 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr9_+_44721808 | 0.38 |
ENSDART00000190578
|
nckap1
|
NCK-associated protein 1 |
| chr11_+_37178271 | 0.37 |
ENSDART00000161771
|
itih3b
|
inter-alpha-trypsin inhibitor heavy chain 3b |
| chr1_+_54137089 | 0.37 |
ENSDART00000062945
|
LO017798.1
|
|
| chr7_-_24005268 | 0.37 |
ENSDART00000173608
|
si:dkey-183c6.9
|
si:dkey-183c6.9 |
| chr11_-_28911172 | 0.37 |
ENSDART00000168493
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr4_+_14900042 | 0.37 |
ENSDART00000018261
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr16_+_1353894 | 0.36 |
ENSDART00000148426
|
celf3b
|
cugbp, Elav-like family member 3b |
| chr15_-_19051152 | 0.36 |
ENSDART00000186453
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr3_-_34296361 | 0.34 |
ENSDART00000181485
ENSDART00000113726 |
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr10_-_41157135 | 0.34 |
ENSDART00000134851
|
aak1b
|
AP2 associated kinase 1b |
| chr1_-_55785722 | 0.32 |
ENSDART00000142069
ENSDART00000043933 |
ndufb7
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 |
| chr22_-_14739491 | 0.31 |
ENSDART00000133385
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr9_-_2572790 | 0.31 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr5_+_65493699 | 0.31 |
ENSDART00000161567
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr22_-_15562933 | 0.31 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr6_-_55347616 | 0.30 |
ENSDART00000172081
|
pcif1
|
PDX1 C-terminal inhibiting factor 1 |
| chr21_-_5205617 | 0.30 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr8_+_3426522 | 0.30 |
ENSDART00000159466
|
CU929199.1
|
|
| chr7_+_6990510 | 0.29 |
ENSDART00000138948
|
ccs
|
copper chaperone for superoxide dismutase |
| chr8_-_1838315 | 0.29 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr20_-_2134620 | 0.28 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr23_-_35064785 | 0.28 |
ENSDART00000172240
|
BX294434.1
|
|
| chr23_-_14990865 | 0.28 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr12_+_10115964 | 0.27 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr16_+_41570653 | 0.26 |
ENSDART00000102665
|
aste1a
|
asteroid homolog 1a |
| chr13_+_23132666 | 0.26 |
ENSDART00000164639
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr9_+_20780813 | 0.26 |
ENSDART00000142787
|
fam46c
|
family with sequence similarity 46, member C |
| chr12_+_5081759 | 0.25 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr1_+_35194454 | 0.24 |
ENSDART00000165389
|
sc:d189
|
sc:d189 |
| chr17_+_5915875 | 0.24 |
ENSDART00000184179
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr13_+_50151407 | 0.24 |
ENSDART00000031858
|
gpr137ba
|
G protein-coupled receptor 137Ba |
| chr11_+_29856032 | 0.24 |
ENSDART00000079150
|
grpr
|
gastrin-releasing peptide receptor |
| chr8_-_26792912 | 0.24 |
ENSDART00000139787
|
kazna
|
kazrin, periplakin interacting protein a |
| chr23_-_44903048 | 0.23 |
ENSDART00000149103
|
fhdc5
|
FH2 domain containing 5 |
| chr24_-_31843173 | 0.22 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr11_+_2687395 | 0.22 |
ENSDART00000082510
|
b3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 |
| chr3_-_44059902 | 0.22 |
ENSDART00000158485
ENSDART00000159088 ENSDART00000165628 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr6_+_54239625 | 0.21 |
ENSDART00000149542
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr2_+_6127593 | 0.21 |
ENSDART00000184007
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr6_-_15641686 | 0.21 |
ENSDART00000135583
|
mlpha
|
melanophilin a |
| chr22_+_30543437 | 0.20 |
ENSDART00000137983
|
si:dkey-103k4.1
|
si:dkey-103k4.1 |
| chr22_-_18164835 | 0.19 |
ENSDART00000143189
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr23_-_41965557 | 0.19 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr19_+_20540843 | 0.19 |
ENSDART00000192500
|
kcnh8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
| chr10_+_6013076 | 0.19 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr12_+_38929663 | 0.18 |
ENSDART00000156334
|
si:dkey-239b22.1
|
si:dkey-239b22.1 |
| chr21_+_38002879 | 0.18 |
ENSDART00000065183
|
cldn2
|
claudin 2 |
| chr4_+_78058635 | 0.18 |
ENSDART00000162631
ENSDART00000191619 |
CABZ01075274.1
|
|
| chr23_-_32092443 | 0.17 |
ENSDART00000133688
|
letmd1
|
LETM1 domain containing 1 |
| chr12_-_2993095 | 0.17 |
ENSDART00000152316
|
si:dkey-202c14.3
|
si:dkey-202c14.3 |
| chr13_-_40238813 | 0.17 |
ENSDART00000044963
|
loxl4
|
lysyl oxidase-like 4 |
| chr1_-_493218 | 0.17 |
ENSDART00000031635
|
ercc5
|
excision repair cross-complementation group 5 |
| chr25_-_24248000 | 0.16 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr13_+_28747934 | 0.16 |
ENSDART00000160176
ENSDART00000101633 ENSDART00000136318 |
prom2
|
prominin 2 |
| chr12_-_3992839 | 0.16 |
ENSDART00000126100
|
si:ch211-136m16.8
|
si:ch211-136m16.8 |
| chr14_-_12307522 | 0.16 |
ENSDART00000163900
|
myot
|
myotilin |
| chr21_-_35082715 | 0.16 |
ENSDART00000146454
|
adrb2b
|
adrenoceptor beta 2, surface b |
| chr23_-_27692717 | 0.15 |
ENSDART00000053878
ENSDART00000145028 |
IKZF4
|
si:dkey-166n8.9 |
| chr24_-_3783497 | 0.15 |
ENSDART00000158354
|
adarb2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr19_+_2279051 | 0.15 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr15_-_4094888 | 0.15 |
ENSDART00000166307
|
TM4SF19
|
si:dkey-83h2.3 |
| chr14_-_899170 | 0.14 |
ENSDART00000165211
ENSDART00000031992 |
rgs14a
|
regulator of G protein signaling 14a |
| chr14_+_23874062 | 0.14 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr7_+_33145529 | 0.14 |
ENSDART00000052387
|
itln2
|
intelectin 2 |
| chr2_+_42871831 | 0.14 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr1_+_12068800 | 0.14 |
ENSDART00000055566
|
xpa
|
xeroderma pigmentosum, complementation group A |
| chr14_-_16379843 | 0.13 |
ENSDART00000090711
|
ltc4s
|
leukotriene C4 synthase |
| chr11_+_44804685 | 0.13 |
ENSDART00000163660
|
strn
|
striatin, calmodulin binding protein |
| chr20_+_38031439 | 0.13 |
ENSDART00000153208
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr22_-_547748 | 0.13 |
ENSDART00000037455
ENSDART00000140101 |
ccnd3
|
cyclin D3 |
| chr18_-_7539469 | 0.13 |
ENSDART00000101296
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr14_+_1007169 | 0.12 |
ENSDART00000172676
|
f8
|
coagulation factor VIII, procoagulant component |
| chr5_+_24882633 | 0.12 |
ENSDART00000111302
|
rhbdd3
|
rhomboid domain containing 3 |
| chr15_+_5339056 | 0.11 |
ENSDART00000174319
|
CABZ01018874.2
|
|
| chr19_+_22727940 | 0.11 |
ENSDART00000052509
|
trhrb
|
thyrotropin-releasing hormone receptor b |
| chr14_-_3132188 | 0.10 |
ENSDART00000172259
|
si:zfos-1069f5.1
|
si:zfos-1069f5.1 |
| chr22_+_1887750 | 0.10 |
ENSDART00000180917
|
si:dkey-15h8.17
|
si:dkey-15h8.17 |
| chr22_+_31295791 | 0.10 |
ENSDART00000092447
|
grip2b
|
glutamate receptor interacting protein 2b |
| chr5_+_1983495 | 0.10 |
ENSDART00000179860
|
CT573103.2
|
|
| chr11_+_3254524 | 0.10 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr3_-_13461361 | 0.10 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr9_+_41156818 | 0.10 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr5_+_13427826 | 0.10 |
ENSDART00000083359
|
sec14l8
|
SEC14-like lipid binding 8 |
| chr13_-_4018888 | 0.09 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr3_+_18398876 | 0.09 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr15_-_5252673 | 0.09 |
ENSDART00000132963
|
or126-1
|
odorant receptor, family E, subfamily 126, member 1 |
| chr19_-_3255725 | 0.08 |
ENSDART00000105165
|
illr1
|
immune-related, lectin-like receptor 1 |
| chr16_-_50897887 | 0.08 |
ENSDART00000156985
|
si:ch73-90p23.1
|
si:ch73-90p23.1 |
| chr22_+_28818291 | 0.07 |
ENSDART00000136032
|
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr10_-_16065185 | 0.07 |
ENSDART00000187266
|
si:dkey-184a18.5
|
si:dkey-184a18.5 |
| chr25_-_28926631 | 0.07 |
ENSDART00000112850
|
etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr25_+_10909850 | 0.07 |
ENSDART00000186021
|
CR339041.3
|
|
| chr10_-_26430168 | 0.07 |
ENSDART00000128894
|
dchs1b
|
dachsous cadherin-related 1b |
| chr14_+_23520986 | 0.07 |
ENSDART00000170473
ENSDART00000175970 |
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr22_-_16755885 | 0.07 |
ENSDART00000036467
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr10_-_5016997 | 0.06 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr24_-_26328721 | 0.06 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr5_-_36674139 | 0.06 |
ENSDART00000188100
|
BX005407.1
|
|
| chr10_+_9697097 | 0.06 |
ENSDART00000167821
|
rabgap1
|
RAB GTPase activating protein 1 |
| chr11_-_45152702 | 0.06 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr15_-_19669770 | 0.06 |
ENSDART00000152707
|
si:dkey-4p15.3
|
si:dkey-4p15.3 |
| chr3_-_33427803 | 0.06 |
ENSDART00000075495
|
rpl23
|
ribosomal protein L23 |
| chr23_-_27015160 | 0.05 |
ENSDART00000189028
|
CU862020.1
|
|
| chr5_-_66768121 | 0.05 |
ENSDART00000141095
|
FERMT3 (1 of many)
|
im:7154036 |
| chr3_+_25195966 | 0.05 |
ENSDART00000112338
|
il2rb
|
interleukin 2 receptor, beta |
| chr22_-_18164671 | 0.04 |
ENSDART00000014057
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr7_-_59210882 | 0.04 |
ENSDART00000170330
ENSDART00000158996 |
nagk
|
N-acetylglucosamine kinase |
| chr15_-_28082310 | 0.03 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr25_+_31405266 | 0.03 |
ENSDART00000103395
|
tnnt3a
|
troponin T type 3a (skeletal, fast) |
| chr10_+_20392083 | 0.03 |
ENSDART00000166867
|
r3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr3_-_39305291 | 0.03 |
ENSDART00000102674
|
plcd3a
|
phospholipase C, delta 3a |
| chr10_+_8690936 | 0.02 |
ENSDART00000144218
|
si:dkey-27b3.4
|
si:dkey-27b3.4 |
| chr15_+_2190229 | 0.02 |
ENSDART00000147710
|
rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr4_-_5455506 | 0.02 |
ENSDART00000156593
ENSDART00000154676 |
si:dkey-14d8.22
|
si:dkey-14d8.22 |
| chr3_-_31619463 | 0.01 |
ENSDART00000124559
|
moto
|
minamoto |
| chr25_-_18953322 | 0.01 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr10_-_18463934 | 0.01 |
ENSDART00000133116
ENSDART00000113422 |
si:dkey-28o19.1
|
si:dkey-28o19.1 |
| chr21_+_19062124 | 0.01 |
ENSDART00000134746
|
rpl17
|
ribosomal protein L17 |
| chr20_+_36628059 | 0.01 |
ENSDART00000062898
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr18_+_7073130 | 0.01 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr16_-_21917361 | 0.00 |
ENSDART00000171818
|
setdb1b
|
SET domain, bifurcated 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of msx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 0.7 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 0.5 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.6 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.1 | 0.4 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 0.2 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.6 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 0.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.2 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 0.2 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.1 | 0.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 0.9 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.0 | 0.1 | GO:1901255 | UV-damage excision repair(GO:0070914) nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.0 | 0.3 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.5 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 1.4 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.1 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0071451 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.1 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.3 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.4 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.0 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.5 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0034354 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.1 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.4 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 1.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.6 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.6 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.3 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.6 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |