Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
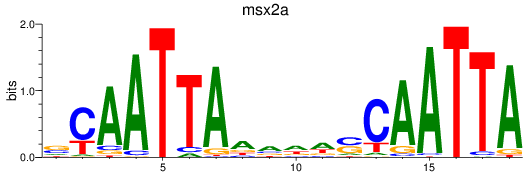
Results for msx2a
Z-value: 0.94
Transcription factors associated with msx2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msx2a
|
ENSDARG00000104651 | muscle segment homeobox 2a |
|
msx2a
|
ENSDARG00000115813 | muscle segment homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msx2a | dr11_v1_chr14_-_24081929_24081929 | 0.37 | 1.2e-01 | Click! |
Activity profile of msx2a motif
Sorted Z-values of msx2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_7666021 | 2.94 |
ENSDART00000180007
|
CABZ01021592.1
|
|
| chr5_-_63515210 | 1.92 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr13_-_6081803 | 1.53 |
ENSDART00000099224
|
dld
|
deltaD |
| chr9_-_32753535 | 1.48 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr16_-_55028740 | 1.37 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr3_-_50954607 | 1.29 |
ENSDART00000163810
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr1_-_45215343 | 1.20 |
ENSDART00000014727
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr25_+_14507567 | 1.19 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr9_+_53337974 | 1.19 |
ENSDART00000145138
|
dct
|
dopachrome tautomerase |
| chr21_-_16113477 | 1.13 |
ENSDART00000147588
|
cyb561a3b
|
cytochrome b561 family, member A3b |
| chr16_-_46578523 | 1.10 |
ENSDART00000131061
|
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr4_-_77260727 | 1.09 |
ENSDART00000075770
|
zgc:162948
|
zgc:162948 |
| chr20_-_9436521 | 1.09 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr10_-_29900546 | 1.05 |
ENSDART00000147441
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr3_-_30941362 | 1.04 |
ENSDART00000076830
|
coro1a
|
coronin, actin binding protein, 1A |
| chr3_-_35541378 | 1.03 |
ENSDART00000183075
ENSDART00000022147 |
ndufab1b
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1b |
| chr4_-_23908802 | 0.96 |
ENSDART00000138873
|
celf2
|
cugbp, Elav-like family member 2 |
| chr19_-_868187 | 0.96 |
ENSDART00000186626
|
eomesa
|
eomesodermin homolog a |
| chr21_-_929448 | 0.89 |
ENSDART00000133976
|
txnl1
|
thioredoxin-like 1 |
| chr10_-_1788376 | 0.87 |
ENSDART00000123842
|
epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr21_-_929293 | 0.80 |
ENSDART00000006419
|
txnl1
|
thioredoxin-like 1 |
| chr6_+_18359306 | 0.77 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr8_+_28358161 | 0.77 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr16_+_28578648 | 0.76 |
ENSDART00000149566
|
nmt2
|
N-myristoyltransferase 2 |
| chr1_-_38756870 | 0.75 |
ENSDART00000130324
ENSDART00000148404 |
gpm6ab
|
glycoprotein M6Ab |
| chr7_+_39664055 | 0.72 |
ENSDART00000146171
|
zgc:158564
|
zgc:158564 |
| chr21_+_17005737 | 0.66 |
ENSDART00000101246
|
vps29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr17_-_11329959 | 0.66 |
ENSDART00000015418
|
irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr22_+_37631234 | 0.66 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr25_+_20119466 | 0.65 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr19_-_5103141 | 0.65 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr5_+_56268436 | 0.65 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr18_-_17485419 | 0.65 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr25_+_19095231 | 0.64 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr2_+_54641644 | 0.64 |
ENSDART00000027313
|
ndufv2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2 |
| chr22_+_12366516 | 0.64 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr12_+_47663419 | 0.64 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr5_+_22177033 | 0.63 |
ENSDART00000131223
|
fdx1b
|
ferredoxin 1b |
| chr6_+_52804267 | 0.63 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr8_-_2230128 | 0.62 |
ENSDART00000140427
|
si:dkeyp-117b11.2
|
si:dkeyp-117b11.2 |
| chr3_-_15496295 | 0.62 |
ENSDART00000144369
|
sgf29
|
SAGA complex associated factor 29 |
| chr7_+_59169081 | 0.61 |
ENSDART00000167980
|
ostc
|
oligosaccharyltransferase complex subunit |
| chr8_+_28452738 | 0.60 |
ENSDART00000062706
|
tmem189
|
transmembrane protein 189 |
| chr3_-_26805455 | 0.60 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr20_+_45893173 | 0.58 |
ENSDART00000131169
|
bmp2b
|
bone morphogenetic protein 2b |
| chr1_+_10051763 | 0.58 |
ENSDART00000011701
|
fgb
|
fibrinogen beta chain |
| chr10_+_20128267 | 0.57 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr25_+_20089986 | 0.56 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr4_-_6459863 | 0.56 |
ENSDART00000138367
|
foxp2
|
forkhead box P2 |
| chr11_+_31323746 | 0.56 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr12_-_19007834 | 0.56 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr22_-_24818066 | 0.56 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr20_+_38032143 | 0.55 |
ENSDART00000032161
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr1_+_35473397 | 0.54 |
ENSDART00000158799
|
usp38
|
ubiquitin specific peptidase 38 |
| chr7_+_34688527 | 0.54 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr1_+_35473219 | 0.53 |
ENSDART00000109678
ENSDART00000181635 |
usp38
|
ubiquitin specific peptidase 38 |
| chr20_-_26822522 | 0.51 |
ENSDART00000146326
ENSDART00000046764 ENSDART00000103234 ENSDART00000143267 |
gmds
|
GDP-mannose 4,6-dehydratase |
| chr24_+_9881219 | 0.51 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr20_+_38031439 | 0.50 |
ENSDART00000153208
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr5_-_33039670 | 0.50 |
ENSDART00000141361
|
glipr2
|
GLI pathogenesis-related 2 |
| chr7_+_15872357 | 0.50 |
ENSDART00000165757
|
pax6b
|
paired box 6b |
| chr14_+_46313396 | 0.50 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr10_-_3138403 | 0.49 |
ENSDART00000183365
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr19_+_43604643 | 0.49 |
ENSDART00000151168
|
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr4_-_6416414 | 0.49 |
ENSDART00000191136
|
MDFIC
|
si:ch73-156e19.1 |
| chr10_-_40397354 | 0.48 |
ENSDART00000150775
ENSDART00000156420 |
taar20o
|
trace amine associated receptor 20o |
| chr11_-_16152105 | 0.48 |
ENSDART00000081062
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr25_-_21492630 | 0.48 |
ENSDART00000141481
|
immp2l
|
inner mitochondrial membrane peptidase subunit 2 |
| chr10_+_40690411 | 0.48 |
ENSDART00000146865
|
taar19m
|
trace amine associated receptor 19m |
| chr17_-_31579715 | 0.47 |
ENSDART00000110167
ENSDART00000191092 |
rpap1
|
RNA polymerase II associated protein 1 |
| chr3_-_15496551 | 0.47 |
ENSDART00000124063
ENSDART00000007726 |
sgf29
|
SAGA complex associated factor 29 |
| chr7_-_51773166 | 0.46 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr10_-_5847655 | 0.44 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr19_+_16016038 | 0.44 |
ENSDART00000131319
|
ctps1a
|
CTP synthase 1a |
| chr10_-_40429747 | 0.43 |
ENSDART00000150613
|
taar20r
|
trace amine associated receptor 20r |
| chr22_-_28668442 | 0.43 |
ENSDART00000182377
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr4_+_2267641 | 0.42 |
ENSDART00000165503
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr17_+_43032529 | 0.42 |
ENSDART00000055611
ENSDART00000154863 |
isca2
|
iron-sulfur cluster assembly 2 |
| chr8_-_53535262 | 0.42 |
ENSDART00000167839
ENSDART00000157521 |
actr8
|
ARP8 actin related protein 8 homolog |
| chr16_+_32238020 | 0.42 |
ENSDART00000017562
|
klhl32
|
kelch-like family member 32 |
| chr24_-_20926812 | 0.42 |
ENSDART00000130958
|
ccdc58
|
coiled-coil domain containing 58 |
| chr22_+_30447969 | 0.41 |
ENSDART00000193066
|
si:dkey-169i5.4
|
si:dkey-169i5.4 |
| chr3_-_26806032 | 0.41 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr4_+_17844013 | 0.41 |
ENSDART00000019165
|
apaf1
|
apoptotic peptidase activating factor 1 |
| chr10_+_40606084 | 0.40 |
ENSDART00000133119
|
si:ch211-238p8.24
|
si:ch211-238p8.24 |
| chr12_-_6063328 | 0.39 |
ENSDART00000002583
|
aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr10_+_40660772 | 0.39 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr19_+_43604256 | 0.39 |
ENSDART00000151080
ENSDART00000110305 |
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr4_+_19700308 | 0.39 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr1_-_22678471 | 0.38 |
ENSDART00000128918
|
fgfbp1b
|
fibroblast growth factor binding protein 1b |
| chr21_+_5192016 | 0.38 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr6_+_30430591 | 0.38 |
ENSDART00000108943
|
shroom2a
|
shroom family member 2a |
| chr10_-_40503395 | 0.38 |
ENSDART00000134499
ENSDART00000193101 |
si:dkey-181m19.17
|
si:dkey-181m19.17 |
| chr11_+_5468629 | 0.37 |
ENSDART00000013203
|
cse1l
|
CSE1 chromosome segregation 1-like (yeast) |
| chr20_-_38836161 | 0.37 |
ENSDART00000061358
|
si:dkey-221h15.4
|
si:dkey-221h15.4 |
| chr6_+_25257728 | 0.37 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr1_-_35924495 | 0.37 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr3_+_28502419 | 0.37 |
ENSDART00000151081
|
sept12
|
septin 12 |
| chr7_+_4474880 | 0.36 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr7_+_27603211 | 0.36 |
ENSDART00000148782
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr21_-_39639954 | 0.35 |
ENSDART00000026766
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr3_+_32410746 | 0.35 |
ENSDART00000025496
|
rras
|
RAS related |
| chr19_-_14970813 | 0.35 |
ENSDART00000184312
|
CR394532.2
|
|
| chr12_+_6098713 | 0.35 |
ENSDART00000139054
|
sgms1
|
sphingomyelin synthase 1 |
| chr4_+_2252123 | 0.35 |
ENSDART00000163996
ENSDART00000066491 |
glipr1a
|
GLI pathogenesis-related 1a |
| chr20_+_18325556 | 0.35 |
ENSDART00000123559
|
znf521
|
zinc finger protein 521 |
| chr19_-_31584444 | 0.35 |
ENSDART00000052183
|
zgc:111986
|
zgc:111986 |
| chr10_-_40366360 | 0.35 |
ENSDART00000188830
ENSDART00000192043 ENSDART00000150415 |
taar20i
|
trace amine associated receptor 20i |
| chr4_-_25271455 | 0.34 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr16_+_28578352 | 0.34 |
ENSDART00000149306
|
nmt2
|
N-myristoyltransferase 2 |
| chr11_+_31324335 | 0.33 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr7_+_69653981 | 0.33 |
ENSDART00000090165
|
frem1a
|
Fras1 related extracellular matrix 1a |
| chr3_+_59880317 | 0.33 |
ENSDART00000166922
ENSDART00000108647 |
alyref
|
Aly/REF export factor |
| chr13_-_17723417 | 0.33 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr16_-_26255877 | 0.32 |
ENSDART00000146214
|
erfl1
|
Ets2 repressor factor like 1 |
| chr10_+_40756352 | 0.32 |
ENSDART00000156210
ENSDART00000144576 |
taar19f
|
trace amine associated receptor 19f |
| chr17_+_45454943 | 0.32 |
ENSDART00000074838
|
kcnk3b
|
potassium channel, subfamily K, member 3b |
| chr10_-_3138859 | 0.31 |
ENSDART00000190606
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr13_+_18321140 | 0.31 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr10_-_5847904 | 0.30 |
ENSDART00000161096
|
ankrd55
|
ankyrin repeat domain 55 |
| chr2_+_36701322 | 0.30 |
ENSDART00000002510
|
golim4b
|
golgi integral membrane protein 4b |
| chr25_-_35101673 | 0.30 |
ENSDART00000140864
|
zgc:162611
|
zgc:162611 |
| chr10_-_20524592 | 0.30 |
ENSDART00000185048
|
ddhd2
|
DDHD domain containing 2 |
| chr20_+_26556372 | 0.29 |
ENSDART00000187179
ENSDART00000157291 |
irf4b
|
interferon regulatory factor 4b |
| chr6_-_32045951 | 0.29 |
ENSDART00000016629
ENSDART00000139055 |
efcab7
|
EF-hand calcium binding domain 7 |
| chr10_+_40568735 | 0.28 |
ENSDART00000136468
ENSDART00000182841 |
taar18i
|
trace amine associated receptor 18i |
| chr17_+_41992054 | 0.28 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr10_+_40633990 | 0.28 |
ENSDART00000190489
ENSDART00000139474 |
si:ch211-238p8.31
|
si:ch211-238p8.31 |
| chr19_-_7441948 | 0.28 |
ENSDART00000003544
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr11_+_17984354 | 0.27 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr23_+_19813677 | 0.27 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr5_+_36752943 | 0.27 |
ENSDART00000017138
|
exoc3l2a
|
exocyst complex component 3-like 2a |
| chr25_-_35101396 | 0.27 |
ENSDART00000138865
|
zgc:162611
|
zgc:162611 |
| chr10_-_40448736 | 0.26 |
ENSDART00000137644
ENSDART00000168190 |
taar20p
|
trace amine associated receptor 20p |
| chr5_+_12528693 | 0.26 |
ENSDART00000051670
|
rfc5
|
replication factor C (activator 1) 5 |
| chr11_+_24046179 | 0.26 |
ENSDART00000006703
|
maf1
|
MAF1 homolog, negative regulator of RNA polymerase III |
| chr14_-_4076480 | 0.26 |
ENSDART00000059231
|
enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr13_-_31008275 | 0.25 |
ENSDART00000139394
|
wdfy4
|
WDFY family member 4 |
| chr10_-_40333319 | 0.25 |
ENSDART00000150479
|
taar20a
|
trace amine associated receptor 20a |
| chr10_+_40774215 | 0.25 |
ENSDART00000131493
|
taar19b
|
trace amine associated receptor 19b |
| chr1_-_17569793 | 0.25 |
ENSDART00000125125
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr10_-_40514643 | 0.24 |
ENSDART00000140705
|
taar19k
|
trace amine associated receptor 19k |
| chr7_-_26408472 | 0.24 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr12_-_29305533 | 0.24 |
ENSDART00000189410
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr3_-_25492361 | 0.24 |
ENSDART00000147322
ENSDART00000055473 |
grb2b
|
growth factor receptor-bound protein 2b |
| chr10_+_40731834 | 0.23 |
ENSDART00000182900
ENSDART00000139284 |
taar19i
|
trace amine associated receptor 19i |
| chr8_+_41048501 | 0.23 |
ENSDART00000123288
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr10_+_40665182 | 0.23 |
ENSDART00000099156
|
taar19u
|
trace amine associated receptor 19u |
| chr2_-_57837838 | 0.23 |
ENSDART00000010699
|
sf3a2
|
splicing factor 3a, subunit 2 |
| chr3_+_48206469 | 0.22 |
ENSDART00000161251
|
si:ch211-207b24.4
|
si:ch211-207b24.4 |
| chr10_-_40484531 | 0.22 |
ENSDART00000142464
ENSDART00000180318 |
taar20z
|
trace amine associated receptor 20z |
| chr19_-_24224142 | 0.22 |
ENSDART00000136409
ENSDART00000114390 |
prf1.8
|
perforin 1.8 |
| chr15_+_2520319 | 0.21 |
ENSDART00000063329
|
cux1b
|
cut-like homeobox 1b |
| chr10_+_20590190 | 0.21 |
ENSDART00000131819
|
letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr22_+_26853254 | 0.21 |
ENSDART00000182487
|
tmem186
|
transmembrane protein 186 |
| chr7_+_34687969 | 0.21 |
ENSDART00000182013
ENSDART00000188255 ENSDART00000182728 |
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr10_+_40629616 | 0.21 |
ENSDART00000147476
|
CR396590.9
|
|
| chr10_+_40768203 | 0.20 |
ENSDART00000171994
ENSDART00000140343 |
taar19d
|
trace amine associated receptor 19d |
| chr10_-_40490647 | 0.20 |
ENSDART00000143660
|
taar20x
|
trace amine associated receptor 20x |
| chr7_-_7493758 | 0.20 |
ENSDART00000036703
|
pfdn2
|
prefoldin subunit 2 |
| chr16_+_10918252 | 0.20 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr10_-_40360635 | 0.20 |
ENSDART00000131970
|
taar20g
|
trace amine associated receptor 20g |
| chr24_+_14713776 | 0.20 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr4_-_14192254 | 0.20 |
ENSDART00000143804
|
pus7l
|
pseudouridylate synthase 7-like |
| chr10_+_40684758 | 0.19 |
ENSDART00000133648
|
taar19h
|
trace amine associated receptor 19h |
| chr10_-_40479911 | 0.19 |
ENSDART00000136741
|
taar20d1
|
trace amine associated receptor 20d1 |
| chr4_+_5796761 | 0.19 |
ENSDART00000164854
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr2_-_10192459 | 0.19 |
ENSDART00000128535
ENSDART00000017173 |
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr10_-_40443722 | 0.19 |
ENSDART00000132906
ENSDART00000157598 |
taar20q
|
trace amine associated receptor 20q |
| chr7_+_13918349 | 0.18 |
ENSDART00000172772
ENSDART00000173384 |
si:cabz01059983.1
|
si:cabz01059983.1 |
| chr6_-_7123210 | 0.18 |
ENSDART00000041304
|
atg3
|
autophagy related 3 |
| chr16_-_28878080 | 0.18 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr15_+_3825117 | 0.18 |
ENSDART00000183315
|
CABZ01061591.1
|
|
| chr8_-_16788626 | 0.17 |
ENSDART00000191652
|
CR759968.2
|
|
| chr19_-_11315224 | 0.17 |
ENSDART00000104933
|
eepd1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr10_+_40726399 | 0.17 |
ENSDART00000142812
|
taar19j
|
trace amine associated receptor 19j |
| chr10_+_40781052 | 0.17 |
ENSDART00000191061
ENSDART00000134435 |
taar19c
|
trace amine associated receptor 19c |
| chr10_+_20589969 | 0.16 |
ENSDART00000183042
|
letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr10_+_40742685 | 0.16 |
ENSDART00000184858
ENSDART00000140300 |
taar19e
|
trace amine associated receptor 19e |
| chr6_-_16717878 | 0.16 |
ENSDART00000153552
|
nomo
|
nodal modulator |
| chr25_-_18739924 | 0.16 |
ENSDART00000156328
|
si:dkeyp-93a5.2
|
si:dkeyp-93a5.2 |
| chr11_+_17984167 | 0.16 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr4_-_9196291 | 0.16 |
ENSDART00000153963
|
hcfc2
|
host cell factor C2 |
| chr10_+_40578332 | 0.15 |
ENSDART00000146155
|
si:ch211-238p8.18
|
si:ch211-238p8.18 |
| chr4_+_77907740 | 0.15 |
ENSDART00000172216
|
si:zfos-2131b9.2
|
si:zfos-2131b9.2 |
| chr7_+_34790768 | 0.15 |
ENSDART00000075116
|
si:dkey-148a17.6
|
si:dkey-148a17.6 |
| chr10_+_40737540 | 0.15 |
ENSDART00000125577
|
taar19a
|
trace amine associated receptor 19a |
| chr6_+_8315050 | 0.15 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr10_-_40454685 | 0.14 |
ENSDART00000137832
|
taar20o
|
trace amine associated receptor 20o |
| chr15_+_8767650 | 0.14 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr25_-_37248795 | 0.14 |
ENSDART00000087247
ENSDART00000154045 |
glg1a
|
golgi glycoprotein 1a |
| chr24_+_20927135 | 0.14 |
ENSDART00000144883
ENSDART00000131829 |
fam162a
|
family with sequence similarity 162, member A |
| chr10_-_40508527 | 0.14 |
ENSDART00000121514
ENSDART00000133729 |
taar20b1
|
trace amine associated receptor 20b1 |
| chr7_-_47282853 | 0.13 |
ENSDART00000169299
|
si:ch211-186j3.3
|
si:ch211-186j3.3 |
| chr24_+_17269849 | 0.13 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr22_-_9183944 | 0.13 |
ENSDART00000188599
|
si:ch211-213a13.5
|
si:ch211-213a13.5 |
| chr10_-_43844537 | 0.13 |
ENSDART00000114208
|
tlr8b
|
toll-like receptor 8b |
| chr16_-_36748374 | 0.13 |
ENSDART00000133310
|
pik3r4
|
phosphoinositide-3-kinase, regulatory subunit 4 |
| chr15_-_29348212 | 0.13 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr14_+_29941266 | 0.12 |
ENSDART00000112757
|
fam149a
|
family with sequence similarity 149 member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of msx2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 | 1.2 | GO:0055057 | neuroblast division(GO:0055057) positive regulation of stem cell proliferation(GO:2000648) |
| 0.4 | 1.5 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.3 | 1.0 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.3 | 1.1 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.2 | 1.5 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.2 | 0.6 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.2 | 0.5 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 0.6 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.5 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 1.3 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.5 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.1 | 0.6 | GO:0034650 | glucocorticoid biosynthetic process(GO:0006704) cortisol metabolic process(GO:0034650) |
| 0.1 | 0.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.8 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.4 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.5 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 0.5 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.4 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.9 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.4 | GO:0007589 | body fluid secretion(GO:0007589) |
| 0.0 | 1.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.6 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.6 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 1.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.7 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 1.2 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.2 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.1 | GO:0003418 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 1.1 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.4 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 1.1 | GO:0042593 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 0.1 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.0 | 1.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 1.2 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 1.0 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.6 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.6 | GO:0035138 | pectoral fin morphogenesis(GO:0035138) |
| 0.0 | 0.2 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.0 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.2 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.5 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 1.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.5 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.6 | GO:0005861 | troponin complex(GO:0005861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.3 | 1.0 | GO:0000035 | acyl binding(GO:0000035) |
| 0.3 | 1.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.2 | 1.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 0.6 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 1.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.7 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.3 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.1 | 8.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.6 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.1 | 0.4 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 0.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.5 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.2 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 1.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.2 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |