Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
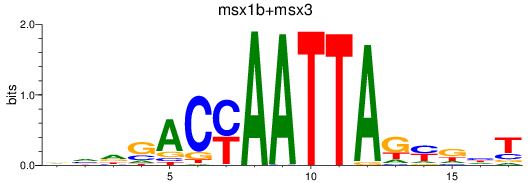
Results for msx1b+msx3
Z-value: 0.77
Transcription factors associated with msx1b+msx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msx1b
|
ENSDARG00000008886 | muscle segment homeobox 1b |
|
msx3
|
ENSDARG00000015674 | muscle segment homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msx1b | dr11_v1_chr1_+_49352900_49352900 | 0.70 | 7.8e-04 | Click! |
| msx3 | dr11_v1_chr13_+_24662238_24662238 | 0.56 | 1.2e-02 | Click! |
Activity profile of msx1b+msx3 motif
Sorted Z-values of msx1b+msx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_23692462 | 1.62 |
ENSDART00000145934
|
hoxb7a
|
homeobox B7a |
| chr21_+_15592426 | 1.22 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr9_-_32343673 | 1.17 |
ENSDART00000078499
|
rftn2
|
raftlin family member 2 |
| chr7_-_8374950 | 1.13 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr20_-_9436521 | 1.11 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr4_-_77260727 | 1.06 |
ENSDART00000075770
|
zgc:162948
|
zgc:162948 |
| chr20_+_11731039 | 1.02 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr14_+_9421510 | 1.02 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr15_-_25518084 | 0.97 |
ENSDART00000158594
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr19_-_19871211 | 0.96 |
ENSDART00000170980
|
evx1
|
even-skipped homeobox 1 |
| chr4_+_19700308 | 0.95 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr17_+_26352372 | 0.91 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr16_-_7379328 | 0.91 |
ENSDART00000060447
|
med18
|
mediator of RNA polymerase II transcription, subunit 18 homolog (yeast) |
| chr3_+_32663865 | 0.89 |
ENSDART00000190240
|
si:dkey-16l2.16
|
si:dkey-16l2.16 |
| chr5_+_29794058 | 0.89 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr13_+_30903816 | 0.88 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr23_-_16485190 | 0.83 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr6_-_9695294 | 0.82 |
ENSDART00000162728
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr21_-_20328375 | 0.82 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr13_+_18321140 | 0.80 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr13_+_9468535 | 0.80 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr24_+_22485710 | 0.78 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr13_-_9213207 | 0.76 |
ENSDART00000139861
ENSDART00000140524 |
HTRA2 (1 of many)
|
si:dkey-33c12.11 |
| chr2_+_6253246 | 0.75 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr22_-_26274177 | 0.74 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr21_-_38730557 | 0.73 |
ENSDART00000150984
ENSDART00000111885 |
taf9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr13_+_15701596 | 0.72 |
ENSDART00000130832
|
trmt61a
|
tRNA methyltransferase 61A |
| chr18_+_9171778 | 0.71 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr25_+_14697247 | 0.69 |
ENSDART00000180747
|
mpped2
|
metallophosphoesterase domain containing 2b |
| chr16_-_41714988 | 0.66 |
ENSDART00000138798
|
cep85
|
centrosomal protein 85 |
| chr15_-_20024205 | 0.65 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr25_+_19666621 | 0.65 |
ENSDART00000112746
|
zgc:171459
|
zgc:171459 |
| chr21_+_723998 | 0.65 |
ENSDART00000160956
|
oaz1b
|
ornithine decarboxylase antizyme 1b |
| chr10_+_36178713 | 0.65 |
ENSDART00000140816
|
or108-3
|
odorant receptor, family D, subfamily 108, member 3 |
| chr15_+_46344655 | 0.65 |
ENSDART00000155893
|
si:ch1073-340i21.2
|
si:ch1073-340i21.2 |
| chr14_+_34490445 | 0.65 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr6_+_39370587 | 0.64 |
ENSDART00000157165
ENSDART00000155079 |
si:dkey-195m11.8
|
si:dkey-195m11.8 |
| chr15_-_37683768 | 0.62 |
ENSDART00000059609
|
si:dkey-117a8.3
|
si:dkey-117a8.3 |
| chr19_-_8768564 | 0.62 |
ENSDART00000170416
|
si:ch73-350k19.1
|
si:ch73-350k19.1 |
| chr10_+_17235370 | 0.61 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr15_+_7057050 | 0.60 |
ENSDART00000061828
|
foxl2a
|
forkhead box L2a |
| chr3_-_31715714 | 0.59 |
ENSDART00000051542
|
ccdc47
|
coiled-coil domain containing 47 |
| chr2_+_16487443 | 0.58 |
ENSDART00000114980
|
CR377211.1
|
|
| chr1_+_55730205 | 0.58 |
ENSDART00000148075
|
adgre10
|
adhesion G protein-coupled receptor E10 |
| chr24_-_7957581 | 0.57 |
ENSDART00000145815
|
txndc5
|
thioredoxin domain containing 5 |
| chr21_+_6751760 | 0.57 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr15_+_36188171 | 0.57 |
ENSDART00000156598
|
masp1
|
mannan-binding lectin serine peptidase 1 |
| chr15_-_37673733 | 0.56 |
ENSDART00000154513
|
si:dkey-117a8.4
|
si:dkey-117a8.4 |
| chr17_-_22021213 | 0.56 |
ENSDART00000078843
|
zmp:0000001102
|
zmp:0000001102 |
| chr16_-_26855936 | 0.55 |
ENSDART00000167320
ENSDART00000078119 |
ino80c
|
INO80 complex subunit C |
| chr24_+_17345521 | 0.54 |
ENSDART00000024722
ENSDART00000154250 |
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr13_-_9045879 | 0.53 |
ENSDART00000155463
ENSDART00000140041 ENSDART00000137454 |
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr8_+_18555559 | 0.53 |
ENSDART00000149523
|
tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr13_+_23282549 | 0.52 |
ENSDART00000101134
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr13_-_9159852 | 0.52 |
ENSDART00000170185
ENSDART00000158697 ENSDART00000143393 ENSDART00000164973 ENSDART00000159910 |
HTRA2 (1 of many)
|
si:dkey-112g5.16 |
| chr19_-_18127629 | 0.52 |
ENSDART00000187722
|
snx10a
|
sorting nexin 10a |
| chr24_-_17039638 | 0.52 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr2_+_30032303 | 0.51 |
ENSDART00000151841
|
rbm33b
|
RNA binding motif protein 33b |
| chr16_+_33144306 | 0.51 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr23_+_44427188 | 0.51 |
ENSDART00000160207
|
eif4e2rs1
|
eukaryotic translation initiation factor 4E family member 2 related sequence 1 |
| chr17_+_41081512 | 0.49 |
ENSDART00000056742
|
babam2
|
BRISC and BRCA1 A complex member 2 |
| chr21_-_37973081 | 0.48 |
ENSDART00000136569
|
ripply1
|
ripply transcriptional repressor 1 |
| chr15_+_15856178 | 0.48 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr3_-_31716157 | 0.47 |
ENSDART00000193189
|
ccdc47
|
coiled-coil domain containing 47 |
| chr6_-_18971063 | 0.46 |
ENSDART00000188163
|
sept9b
|
septin 9b |
| chr17_+_24821627 | 0.45 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr16_+_33144112 | 0.45 |
ENSDART00000183149
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr12_-_33357655 | 0.44 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr24_-_9979342 | 0.44 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr10_+_39212898 | 0.43 |
ENSDART00000159501
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr18_+_48423973 | 0.42 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr14_+_6535426 | 0.41 |
ENSDART00000055961
|
thg1l
|
tRNA-histidine guanylyltransferase 1-like |
| chr5_+_56023186 | 0.41 |
ENSDART00000156230
|
fzd9a
|
frizzled class receptor 9a |
| chr13_+_35955562 | 0.40 |
ENSDART00000137377
|
si:ch211-67f13.7
|
si:ch211-67f13.7 |
| chr15_-_37709072 | 0.40 |
ENSDART00000157115
|
si:dkey-117a8.2
|
si:dkey-117a8.2 |
| chr2_+_50608099 | 0.40 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr15_-_37620167 | 0.40 |
ENSDART00000059582
|
si:ch211-137j23.8
|
si:ch211-137j23.8 |
| chr17_+_41081977 | 0.40 |
ENSDART00000137091
ENSDART00000143714 |
babam2
|
BRISC and BRCA1 A complex member 2 |
| chr13_+_9521629 | 0.39 |
ENSDART00000149870
ENSDART00000137666 |
si:dkey-19p15.4
|
si:dkey-19p15.4 |
| chr5_-_68093169 | 0.38 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr3_+_34140507 | 0.38 |
ENSDART00000131802
|
si:dkey-204f11.64
|
si:dkey-204f11.64 |
| chr19_-_18127808 | 0.37 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr17_+_24851951 | 0.37 |
ENSDART00000180746
|
cx35.4
|
connexin 35.4 |
| chr9_-_35633827 | 0.36 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr10_+_39212601 | 0.36 |
ENSDART00000168778
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr15_-_14212777 | 0.36 |
ENSDART00000165572
|
CR925813.1
|
|
| chr17_-_50071748 | 0.35 |
ENSDART00000075188
|
zgc:113886
|
zgc:113886 |
| chr24_+_26345427 | 0.35 |
ENSDART00000089756
|
lrrc34
|
leucine rich repeat containing 34 |
| chr16_+_41171949 | 0.35 |
ENSDART00000135294
|
nek11
|
NIMA-related kinase 11 |
| chr6_+_52877147 | 0.34 |
ENSDART00000174370
|
or137-8
|
odorant receptor, family H, subfamily 137, member 8 |
| chr21_-_22647695 | 0.34 |
ENSDART00000187553
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr11_-_40457325 | 0.33 |
ENSDART00000128442
|
tnfrsf1b
|
tumor necrosis factor receptor superfamily, member 1B |
| chr25_-_25550938 | 0.32 |
ENSDART00000150412
ENSDART00000103622 |
irf7
|
interferon regulatory factor 7 |
| chr23_+_22658700 | 0.31 |
ENSDART00000192248
|
eno1a
|
enolase 1a, (alpha) |
| chr16_-_35975254 | 0.31 |
ENSDART00000167537
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr20_-_19511016 | 0.30 |
ENSDART00000168521
|
snx17
|
sorting nexin 17 |
| chr24_+_26345609 | 0.29 |
ENSDART00000186844
|
lrrc34
|
leucine rich repeat containing 34 |
| chr15_-_31409013 | 0.28 |
ENSDART00000140456
|
or111-9
|
odorant receptor, family D, subfamily 111, member 9 |
| chr11_-_25538341 | 0.28 |
ENSDART00000171560
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr10_+_11355841 | 0.28 |
ENSDART00000193067
ENSDART00000064215 |
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr10_-_44560165 | 0.26 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr10_+_40568735 | 0.25 |
ENSDART00000136468
ENSDART00000182841 |
taar18i
|
trace amine associated receptor 18i |
| chr19_+_9212031 | 0.25 |
ENSDART00000052930
|
ndufv1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1 |
| chr24_+_38306010 | 0.25 |
ENSDART00000143184
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr22_+_5532003 | 0.24 |
ENSDART00000106174
|
si:ch73-256j6.2
|
si:ch73-256j6.2 |
| chr4_-_12997348 | 0.24 |
ENSDART00000171451
|
si:dkey-6a5.3
|
si:dkey-6a5.3 |
| chr7_+_24496894 | 0.24 |
ENSDART00000149994
|
nelfa
|
negative elongation factor complex member A |
| chr8_-_15129573 | 0.23 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr17_+_7513673 | 0.22 |
ENSDART00000156674
|
klhl10b.1
|
kelch-like family member 10b, tandem duplicate 1 |
| chr9_+_35832294 | 0.22 |
ENSDART00000130549
ENSDART00000122169 |
si:dkey-67c22.2
|
si:dkey-67c22.2 |
| chr4_+_54519511 | 0.22 |
ENSDART00000161653
|
znf974
|
zinc finger protein 974 |
| chr5_+_27897504 | 0.22 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr15_-_37733238 | 0.22 |
ENSDART00000115178
|
si:dkey-42l23.7
|
si:dkey-42l23.7 |
| chr23_-_42876596 | 0.21 |
ENSDART00000086156
|
dlgap4a
|
discs, large (Drosophila) homolog-associated protein 4a |
| chr15_-_37767901 | 0.21 |
ENSDART00000154763
|
si:dkey-42l23.4
|
si:dkey-42l23.4 |
| chr22_+_12770877 | 0.21 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr21_-_22648007 | 0.21 |
ENSDART00000121788
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr5_+_9224051 | 0.20 |
ENSDART00000139265
|
si:ch211-12e13.12
|
si:ch211-12e13.12 |
| chr22_+_9114416 | 0.19 |
ENSDART00000190169
|
nlrp15
|
NACHT, LRR and PYD domains-containing protein 15 |
| chr4_+_77047067 | 0.19 |
ENSDART00000174206
|
znf1009
|
zinc finger protein 1009 |
| chr10_+_36194527 | 0.19 |
ENSDART00000076810
|
or108-1
|
odorant receptor, family D, subfamily 108, member 1 |
| chr19_+_1673599 | 0.19 |
ENSDART00000163127
|
klhl7
|
kelch-like family member 7 |
| chr10_+_40598791 | 0.19 |
ENSDART00000131895
|
taar17a
|
trace amine associated receptor 17a |
| chr19_+_8985230 | 0.18 |
ENSDART00000018973
|
scamp3
|
secretory carrier membrane protein 3 |
| chr23_+_16889352 | 0.18 |
ENSDART00000145275
|
si:dkey-147f3.8
|
si:dkey-147f3.8 |
| chr14_-_470505 | 0.18 |
ENSDART00000067147
|
ANKRD50
|
ankyrin repeat domain 50 |
| chr9_+_18829360 | 0.17 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr24_+_35881124 | 0.17 |
ENSDART00000143015
|
klhl14
|
kelch-like family member 14 |
| chr6_-_19042294 | 0.16 |
ENSDART00000159461
|
si:rp71-81e14.2
|
si:rp71-81e14.2 |
| chr7_+_59677273 | 0.16 |
ENSDART00000039535
ENSDART00000132044 |
trmt44
|
tRNA methyltransferase 44 homolog |
| chr15_-_37719015 | 0.16 |
ENSDART00000154875
|
si:dkey-117a8.1
|
si:dkey-117a8.1 |
| chr16_+_20161805 | 0.15 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr3_+_2719243 | 0.15 |
ENSDART00000189256
|
CR388047.3
|
|
| chr4_+_77943184 | 0.14 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr5_-_29512538 | 0.14 |
ENSDART00000098364
|
ehmt1a
|
euchromatic histone-lysine N-methyltransferase 1a |
| chr4_-_9891874 | 0.14 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr8_-_17822259 | 0.13 |
ENSDART00000063587
|
st6galnac5b
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5b |
| chr21_+_6751405 | 0.13 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr20_+_28803977 | 0.12 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr11_+_31864921 | 0.12 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr10_-_40416054 | 0.12 |
ENSDART00000137947
ENSDART00000143606 |
taar20j
|
trace amine associated receptor 20j |
| chr4_-_20292821 | 0.12 |
ENSDART00000136069
ENSDART00000192504 |
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr8_-_7567815 | 0.12 |
ENSDART00000132536
|
hcfc1b
|
host cell factor C1b |
| chr9_+_12444494 | 0.11 |
ENSDART00000102430
|
tmem41aa
|
transmembrane protein 41aa |
| chr4_+_62576422 | 0.11 |
ENSDART00000180679
|
si:ch211-79g12.2
|
si:ch211-79g12.2 |
| chr20_-_51831816 | 0.11 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr13_+_27328098 | 0.11 |
ENSDART00000037585
|
mb21d1
|
Mab-21 domain containing 1 |
| chr25_+_6471401 | 0.10 |
ENSDART00000132927
|
snx33
|
sorting nexin 33 |
| chr17_-_6618574 | 0.09 |
ENSDART00000184486
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr17_-_41081806 | 0.08 |
ENSDART00000030622
|
rbks
|
ribokinase |
| chr18_-_16801033 | 0.08 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr18_+_44630415 | 0.08 |
ENSDART00000098540
|
bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr22_-_21150845 | 0.08 |
ENSDART00000027345
|
tmem59l
|
transmembrane protein 59-like |
| chr5_+_872299 | 0.07 |
ENSDART00000130042
|
fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr16_+_35595312 | 0.06 |
ENSDART00000170438
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr16_+_9713850 | 0.05 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr25_-_27564205 | 0.04 |
ENSDART00000157319
|
hyal4
|
hyaluronoglucosaminidase 4 |
| chr25_+_34748860 | 0.04 |
ENSDART00000087364
|
wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr8_-_27656765 | 0.03 |
ENSDART00000078491
|
mov10b.2
|
Moloney leukemia virus 10b, tandem duplicate 2 |
| chr15_-_42039192 | 0.03 |
ENSDART00000004338
|
epha4l
|
eph receptor A4, like |
| chr18_+_43891051 | 0.03 |
ENSDART00000024213
|
cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr22_+_2769236 | 0.03 |
ENSDART00000141836
|
si:dkey-20i20.10
|
si:dkey-20i20.10 |
| chr12_+_3078221 | 0.02 |
ENSDART00000148835
ENSDART00000149427 |
sgca
|
sarcoglycan, alpha |
| chr13_+_22476742 | 0.02 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
| chr16_+_5774977 | 0.02 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr2_+_10771787 | 0.01 |
ENSDART00000187782
|
gfi1aa
|
growth factor independent 1A transcription repressor a |
Network of associatons between targets according to the STRING database.
First level regulatory network of msx1b+msx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.2 | 0.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 0.6 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 0.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 1.7 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 0.9 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 2.6 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.9 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.9 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.1 | 0.3 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 0.5 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.4 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 0.2 | GO:0019556 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 1.2 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 1.0 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) post-translational protein modification(GO:0043687) |
| 0.0 | 0.7 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 1.0 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.2 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.3 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.0 | 0.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.7 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.3 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.1 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 1.9 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.2 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.0 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 1.3 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.2 | 1.2 | GO:0035060 | brahma complex(GO:0035060) |
| 0.2 | 0.6 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.7 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 0.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.9 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.5 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 0.8 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 0.7 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.7 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 2.6 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 1.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.4 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.4 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.8 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.7 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.9 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.5 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.9 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) FMN binding(GO:0010181) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.0 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.0 | 2.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |