Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
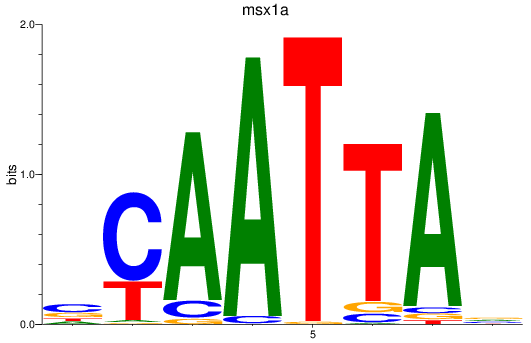
Results for msx1a
Z-value: 1.01
Transcription factors associated with msx1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msx1a
|
ENSDARG00000116118 | muscle segment homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msx1a | dr11_v1_chr14_-_12822_12847 | -0.92 | 3.0e-08 | Click! |
Activity profile of msx1a motif
Sorted Z-values of msx1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_12270857 | 5.03 |
ENSDART00000170093
|
si:dkey-36i7.3
|
si:dkey-36i7.3 |
| chr11_+_30244356 | 4.05 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr3_-_50443607 | 3.88 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr23_+_40460333 | 3.62 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr17_-_12389259 | 3.62 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr25_-_10503043 | 3.56 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr19_+_10339538 | 3.45 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr12_+_24342303 | 3.27 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr2_+_39021282 | 3.00 |
ENSDART00000056577
|
RBP1 (1 of many)
|
si:ch211-119o8.7 |
| chr20_-_40755614 | 2.61 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr13_+_28701233 | 2.49 |
ENSDART00000135931
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr1_-_50859053 | 2.46 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr20_+_30490682 | 2.40 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr25_-_32869794 | 2.36 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr3_-_32169754 | 2.19 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr22_+_35068046 | 2.18 |
ENSDART00000161660
ENSDART00000169573 |
si:ch73-173h19.3
|
si:ch73-173h19.3 |
| chr5_-_23280098 | 2.17 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr12_+_6195191 | 2.15 |
ENSDART00000043236
ENSDART00000186420 |
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr23_-_28141419 | 2.14 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr6_+_3828560 | 2.08 |
ENSDART00000185273
ENSDART00000179091 |
gad1b
|
glutamate decarboxylase 1b |
| chr16_+_5774977 | 2.07 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr4_+_12612723 | 2.05 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr18_+_15644559 | 2.05 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr16_-_29437373 | 1.94 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr15_-_6247775 | 1.91 |
ENSDART00000148350
|
dscamb
|
Down syndrome cell adhesion molecule b |
| chr2_-_33645411 | 1.85 |
ENSDART00000114663
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr8_-_46894362 | 1.85 |
ENSDART00000111124
|
acot7
|
acyl-CoA thioesterase 7 |
| chr18_-_16801033 | 1.84 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr2_+_50626476 | 1.83 |
ENSDART00000018150
|
neurod6b
|
neuronal differentiation 6b |
| chr2_+_31804582 | 1.82 |
ENSDART00000086646
|
rnf182
|
ring finger protein 182 |
| chr20_-_29420713 | 1.76 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr15_+_9053059 | 1.74 |
ENSDART00000012039
|
ppm1na
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Na (putative) |
| chr14_-_2933185 | 1.73 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr24_-_24849091 | 1.69 |
ENSDART00000133649
ENSDART00000038290 |
crhb
|
corticotropin releasing hormone b |
| chr16_-_47483142 | 1.67 |
ENSDART00000147072
|
cthrc1b
|
collagen triple helix repeat containing 1b |
| chr4_+_11384891 | 1.66 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr22_+_28320792 | 1.58 |
ENSDART00000160562
ENSDART00000164138 |
impg2b
impg2b
|
interphotoreceptor matrix proteoglycan 2b interphotoreceptor matrix proteoglycan 2b |
| chr2_+_50608099 | 1.57 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr13_-_44630111 | 1.56 |
ENSDART00000110092
|
mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr23_-_1348933 | 1.53 |
ENSDART00000168981
|
CABZ01078120.1
|
|
| chr20_-_32110882 | 1.53 |
ENSDART00000030324
|
grm1a
|
glutamate receptor, metabotropic 1a |
| chr13_+_38430466 | 1.50 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr22_-_10486477 | 1.49 |
ENSDART00000184366
|
aspn
|
asporin (LRR class 1) |
| chr19_-_8604429 | 1.45 |
ENSDART00000151165
|
trim46b
|
tripartite motif containing 46b |
| chr23_+_28582865 | 1.42 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr19_-_38830582 | 1.41 |
ENSDART00000189966
ENSDART00000183055 |
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr7_+_32722227 | 1.37 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr13_+_25449681 | 1.36 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr21_+_6751405 | 1.34 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr23_+_33963619 | 1.27 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr10_+_42690374 | 1.27 |
ENSDART00000123496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr8_+_22582146 | 1.26 |
ENSDART00000157655
ENSDART00000189892 |
CT583651.2
|
|
| chr13_-_20381485 | 1.25 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr18_+_10884996 | 1.24 |
ENSDART00000147613
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr12_+_2446837 | 1.24 |
ENSDART00000112032
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr1_+_18811679 | 1.23 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr12_+_28749189 | 1.23 |
ENSDART00000013980
|
tbx21
|
T-box 21 |
| chr11_+_12052791 | 1.23 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr22_+_20427170 | 1.21 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr22_-_13649588 | 1.18 |
ENSDART00000131877
|
si:ch211-279g13.1
|
si:ch211-279g13.1 |
| chr14_-_2364761 | 1.14 |
ENSDART00000167322
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.3 |
| chr6_-_14292307 | 1.14 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr10_-_13343831 | 1.13 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr13_-_36911118 | 1.12 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr6_-_6487876 | 1.12 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr22_-_23253481 | 1.12 |
ENSDART00000054807
|
lhx9
|
LIM homeobox 9 |
| chr14_-_30587814 | 1.10 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr16_+_31804590 | 1.07 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr7_-_23996133 | 1.06 |
ENSDART00000173761
|
si:dkey-183c6.8
|
si:dkey-183c6.8 |
| chr18_-_43884044 | 1.06 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr2_-_57076687 | 1.05 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr12_+_31744217 | 1.04 |
ENSDART00000190361
|
RNF157
|
si:dkey-49c17.3 |
| chr22_-_7129631 | 1.04 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr5_+_60928576 | 1.04 |
ENSDART00000131041
|
doc2b
|
double C2-like domains, beta |
| chr22_+_5176693 | 1.03 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr22_+_18477934 | 1.03 |
ENSDART00000132684
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr7_+_30787903 | 1.02 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr10_+_3049636 | 1.02 |
ENSDART00000081794
ENSDART00000183167 ENSDART00000191634 ENSDART00000183514 |
rasgrf2a
|
Ras protein-specific guanine nucleotide-releasing factor 2a |
| chr6_+_23887314 | 1.02 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr11_+_7264457 | 0.99 |
ENSDART00000154182
|
reep6
|
receptor accessory protein 6 |
| chr7_-_28148310 | 0.99 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr20_-_46362606 | 0.99 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr17_-_23616626 | 0.98 |
ENSDART00000104730
|
ifit14
|
interferon-induced protein with tetratricopeptide repeats 14 |
| chr21_+_6751760 | 0.98 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr25_+_19149241 | 0.98 |
ENSDART00000184982
ENSDART00000067324 |
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr1_-_47071979 | 0.97 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr7_-_36096582 | 0.95 |
ENSDART00000188507
|
CR848715.1
|
|
| chr2_+_37227011 | 0.95 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr8_-_20230559 | 0.94 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr16_+_39159752 | 0.94 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr6_-_44711942 | 0.93 |
ENSDART00000055035
|
cntn3b
|
contactin 3b |
| chr5_-_67629263 | 0.89 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr3_-_32337653 | 0.89 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr24_-_31846366 | 0.88 |
ENSDART00000155295
|
steap2
|
STEAP family member 2, metalloreductase |
| chr11_-_42554290 | 0.84 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr2_+_21229909 | 0.81 |
ENSDART00000046127
|
vipr1a
|
vasoactive intestinal peptide receptor 1a |
| chr19_-_42391383 | 0.79 |
ENSDART00000110075
ENSDART00000087002 |
plekho1a
|
pleckstrin homology domain containing, family O member 1a |
| chr6_+_3280939 | 0.79 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr15_-_22074315 | 0.78 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr10_+_16584382 | 0.76 |
ENSDART00000112039
|
CR790388.1
|
|
| chr18_-_2433011 | 0.75 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr15_-_34785594 | 0.71 |
ENSDART00000154256
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr19_+_42470396 | 0.71 |
ENSDART00000191679
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr12_-_20120702 | 0.71 |
ENSDART00000153387
ENSDART00000158412 ENSDART00000112768 |
ubald1a
|
UBA-like domain containing 1a |
| chr6_+_39232245 | 0.68 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr16_-_27628994 | 0.68 |
ENSDART00000157407
|
nacad
|
NAC alpha domain containing |
| chr17_+_33311784 | 0.67 |
ENSDART00000156229
|
si:ch211-132f19.7
|
si:ch211-132f19.7 |
| chr20_-_32270866 | 0.65 |
ENSDART00000153140
|
armc2
|
armadillo repeat containing 2 |
| chr20_-_28800999 | 0.63 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr22_-_10121880 | 0.62 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr20_-_45060241 | 0.62 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr8_+_26125218 | 0.61 |
ENSDART00000145095
|
celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr2_-_23004286 | 0.60 |
ENSDART00000134664
ENSDART00000110373 ENSDART00000185833 ENSDART00000187235 |
znf414
mllt1b
|
zinc finger protein 414 MLLT1, super elongation complex subunit b |
| chr8_+_26253252 | 0.60 |
ENSDART00000142031
|
slc26a6
|
solute carrier family 26, member 6 |
| chr10_+_21867307 | 0.57 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr4_-_15420452 | 0.57 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr21_-_3853204 | 0.56 |
ENSDART00000188829
|
st6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr1_-_36152131 | 0.56 |
ENSDART00000182113
ENSDART00000182904 |
znf827
|
zinc finger protein 827 |
| chr13_+_2625150 | 0.56 |
ENSDART00000164177
|
plpp4
|
phospholipid phosphatase 4 |
| chr25_-_27621268 | 0.54 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr1_-_18811517 | 0.53 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr21_-_8085635 | 0.52 |
ENSDART00000082790
|
si:dkey-163m14.2
|
si:dkey-163m14.2 |
| chr7_-_31941330 | 0.52 |
ENSDART00000144682
|
bdnf
|
brain-derived neurotrophic factor |
| chr22_+_5176255 | 0.52 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr9_+_24008879 | 0.51 |
ENSDART00000190419
ENSDART00000191843 ENSDART00000148226 |
mlphb
|
melanophilin b |
| chr17_+_31914877 | 0.48 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr24_-_4782052 | 0.47 |
ENSDART00000149911
|
agtr1b
|
angiotensin II receptor, type 1b |
| chr11_-_7261717 | 0.45 |
ENSDART00000128959
|
zgc:113223
|
zgc:113223 |
| chr9_+_34331368 | 0.45 |
ENSDART00000147913
|
pou2f1b
|
POU class 2 homeobox 1b |
| chr3_+_56366395 | 0.43 |
ENSDART00000154367
|
cacng5b
|
calcium channel, voltage-dependent, gamma subunit 5b |
| chr14_-_25034142 | 0.42 |
ENSDART00000000392
|
pde6a
|
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| chr3_-_32898626 | 0.42 |
ENSDART00000103201
|
kat7a
|
K(lysine) acetyltransferase 7a |
| chr20_+_19238382 | 0.41 |
ENSDART00000136757
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr25_+_11456696 | 0.40 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr6_+_102506 | 0.39 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr9_-_52962521 | 0.38 |
ENSDART00000170419
|
CU855885.1
|
|
| chr10_-_28513861 | 0.38 |
ENSDART00000177781
|
bbx
|
bobby sox homolog (Drosophila) |
| chr12_+_21298317 | 0.36 |
ENSDART00000178562
|
ca10a
|
carbonic anhydrase Xa |
| chr10_-_34916208 | 0.35 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr6_+_9204099 | 0.34 |
ENSDART00000150167
ENSDART00000115394 |
tbx19
|
T-box 19 |
| chr5_+_11290851 | 0.33 |
ENSDART00000180408
|
CABZ01077124.1
|
|
| chr9_-_21918963 | 0.31 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr15_-_23376541 | 0.31 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr4_-_72638972 | 0.31 |
ENSDART00000193312
|
CABZ01054394.1
|
|
| chr11_-_29768054 | 0.31 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr10_-_32494304 | 0.30 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr19_-_19556778 | 0.29 |
ENSDART00000164060
|
tax1bp1a
|
Tax1 (human T-cell leukemia virus type I) binding protein 1a |
| chr12_-_18872927 | 0.28 |
ENSDART00000187717
|
shisa8b
|
shisa family member 8b |
| chr16_+_54263921 | 0.28 |
ENSDART00000002856
|
drd2l
|
dopamine receptor D2 like |
| chr15_+_23911655 | 0.28 |
ENSDART00000156304
|
si:ch1073-145m9.1
|
si:ch1073-145m9.1 |
| chr24_+_40860320 | 0.24 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr10_-_32494499 | 0.22 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr15_-_8665662 | 0.20 |
ENSDART00000090675
|
arhgap35a
|
Rho GTPase activating protein 35a |
| chr13_+_38990939 | 0.20 |
ENSDART00000145979
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr6_+_11850359 | 0.19 |
ENSDART00000109552
ENSDART00000188139 ENSDART00000181499 ENSDART00000178269 |
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr10_+_43039947 | 0.19 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr15_-_21877726 | 0.18 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr18_+_7204378 | 0.18 |
ENSDART00000142905
|
vwf
|
von Willebrand factor |
| chr1_+_34696503 | 0.18 |
ENSDART00000186106
|
CR339054.2
|
|
| chr7_-_31940590 | 0.17 |
ENSDART00000131009
|
bdnf
|
brain-derived neurotrophic factor |
| chr25_+_10410620 | 0.17 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr15_-_26931541 | 0.16 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr7_-_12464412 | 0.15 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr6_-_39649504 | 0.14 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr22_+_1092479 | 0.14 |
ENSDART00000170119
|
guca1e
|
guanylate cyclase activator 1e |
| chrM_+_11009 | 0.13 |
ENSDART00000093617
|
mt-nd4l
|
NADH dehydrogenase 4L, mitochondrial |
| chr8_-_38477817 | 0.13 |
ENSDART00000075989
|
inpp5l
|
inositol polyphosphate-5-phosphatase L |
| chr4_-_9891874 | 0.13 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr12_+_20352400 | 0.13 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr20_-_29864390 | 0.12 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr16_-_16120941 | 0.12 |
ENSDART00000131227
|
ankib1b
|
ankyrin repeat and IBR domain containing 1b |
| chr21_-_20757866 | 0.11 |
ENSDART00000163003
|
ghrb
|
growth hormone receptor b |
| chr21_-_27338639 | 0.10 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr17_+_25856671 | 0.09 |
ENSDART00000064817
|
wapla
|
WAPL cohesin release factor a |
| chr20_-_11178022 | 0.09 |
ENSDART00000152246
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr5_+_37032038 | 0.08 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr13_-_36184476 | 0.08 |
ENSDART00000057185
|
map3k9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr4_+_16885854 | 0.08 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr10_-_21955231 | 0.08 |
ENSDART00000183695
|
FO744833.2
|
|
| chr21_-_28640316 | 0.08 |
ENSDART00000128237
|
nrg2a
|
neuregulin 2a |
| chr19_+_7735157 | 0.07 |
ENSDART00000186717
|
tuft1b
|
tuftelin 1b |
| chr17_+_26543690 | 0.06 |
ENSDART00000156417
|
sparcl2
|
SPARC-like 2 |
| chr23_+_38957472 | 0.04 |
ENSDART00000193836
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr8_-_25034411 | 0.02 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr8_+_25034544 | 0.01 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr7_-_23768234 | 0.01 |
ENSDART00000173981
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of msx1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.7 | 7.3 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.6 | 1.7 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.5 | 2.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.4 | 1.1 | GO:0097376 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.3 | 1.0 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.3 | 2.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 1.5 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.3 | 3.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 0.9 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 0.9 | GO:0015677 | copper ion import(GO:0015677) |
| 0.2 | 0.6 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.2 | 1.7 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 1.0 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 2.1 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 2.2 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 1.0 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 0.6 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 | 0.6 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 1.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.7 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 1.9 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 1.1 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.5 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 1.7 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 1.9 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.8 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 3.6 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 2.1 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 1.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 2.2 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.3 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 1.0 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 1.4 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 1.7 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.2 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 | 1.1 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 1.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.4 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 2.2 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 1.0 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.2 | GO:0072676 | lymphocyte migration(GO:0072676) |
| 0.0 | 0.7 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.8 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 1.2 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.1 | GO:0060347 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.6 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.3 | 1.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 1.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 1.6 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.2 | 3.6 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 1.7 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 0.8 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 2.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.7 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.7 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 2.0 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 1.1 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.9 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 4.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 1.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.7 | 2.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.4 | 2.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.4 | 1.8 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.3 | 1.0 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.3 | 2.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.3 | 1.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 1.9 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 0.9 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 1.5 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.2 | 1.0 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 0.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 1.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 2.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.8 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 3.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.5 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 4.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 2.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.7 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 1.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.9 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 0.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.2 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.3 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 1.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.7 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 2.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 2.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 1.1 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |