Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
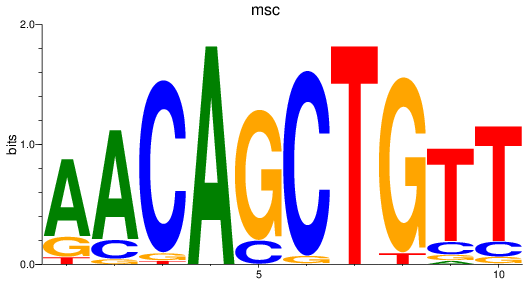
Results for msc
Z-value: 0.52
Transcription factors associated with msc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msc
|
ENSDARG00000110016 | musculin (activated B-cell factor-1) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msc | dr11_v1_chr24_+_13735616_13735616 | -0.34 | 1.5e-01 | Click! |
Activity profile of msc motif
Sorted Z-values of msc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_19400166 | 0.90 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr19_+_1184878 | 0.74 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr2_-_17114852 | 0.71 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr20_-_48485354 | 0.63 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr3_-_21094437 | 0.59 |
ENSDART00000153739
ENSDART00000109790 |
nlk1
|
nemo-like kinase, type 1 |
| chr2_-_17115256 | 0.56 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr21_-_17482465 | 0.51 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr19_-_10214264 | 0.50 |
ENSDART00000053300
ENSDART00000148225 |
znf865
|
zinc finger protein 865 |
| chr16_+_40575742 | 0.48 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr15_-_5624361 | 0.46 |
ENSDART00000176446
ENSDART00000114410 |
wdr62
|
WD repeat domain 62 |
| chr1_+_8694196 | 0.44 |
ENSDART00000025604
|
zgc:77849
|
zgc:77849 |
| chr17_-_14836320 | 0.43 |
ENSDART00000157051
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr23_+_35714574 | 0.43 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr1_-_55118745 | 0.43 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr4_-_21466480 | 0.42 |
ENSDART00000139962
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr8_+_48484455 | 0.42 |
ENSDART00000122737
|
PRDM16
|
si:ch211-263k4.2 |
| chr23_-_27647769 | 0.41 |
ENSDART00000059974
|
wnt10b
|
wingless-type MMTV integration site family, member 10b |
| chr22_+_465269 | 0.41 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr19_-_2317558 | 0.40 |
ENSDART00000190300
|
sp8a
|
sp8 transcription factor a |
| chr1_-_17587552 | 0.40 |
ENSDART00000039917
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr21_+_13205859 | 0.39 |
ENSDART00000102253
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr17_+_23968214 | 0.39 |
ENSDART00000183053
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr14_+_7939216 | 0.39 |
ENSDART00000171657
|
cxxc5b
|
CXXC finger protein 5b |
| chr7_+_6652967 | 0.39 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr12_-_31484677 | 0.38 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr7_+_44715224 | 0.38 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr17_-_51195651 | 0.38 |
ENSDART00000191205
ENSDART00000088185 |
paplna
|
papilin a, proteoglycan-like sulfated glycoprotein |
| chr15_-_32383529 | 0.37 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr8_-_14609284 | 0.37 |
ENSDART00000146175
|
cep350
|
centrosomal protein 350 |
| chr7_+_19552381 | 0.37 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr13_+_43400443 | 0.37 |
ENSDART00000084321
|
dact2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr14_+_7939398 | 0.36 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr13_+_22317216 | 0.36 |
ENSDART00000110794
|
usp54a
|
ubiquitin specific peptidase 54a |
| chr23_+_16948049 | 0.36 |
ENSDART00000139449
|
si:dkey-147f3.4
|
si:dkey-147f3.4 |
| chr18_-_20560007 | 0.36 |
ENSDART00000141367
ENSDART00000090186 |
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr3_-_29962345 | 0.35 |
ENSDART00000136819
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr15_+_32727848 | 0.35 |
ENSDART00000161361
|
postnb
|
periostin, osteoblast specific factor b |
| chr5_+_50879545 | 0.35 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr17_+_38262408 | 0.34 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr19_-_30810328 | 0.34 |
ENSDART00000184875
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr22_-_18179214 | 0.34 |
ENSDART00000129576
|
si:ch211-125m10.6
|
si:ch211-125m10.6 |
| chr14_-_32255126 | 0.32 |
ENSDART00000017259
|
fgf13a
|
fibroblast growth factor 13a |
| chr22_+_24770744 | 0.32 |
ENSDART00000142882
|
si:rp71-23d18.4
|
si:rp71-23d18.4 |
| chr1_+_33401774 | 0.32 |
ENSDART00000149786
ENSDART00000110026 ENSDART00000148946 |
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr11_+_14622379 | 0.31 |
ENSDART00000112589
|
efna2b
|
ephrin-A2b |
| chr18_+_36631923 | 0.31 |
ENSDART00000098980
|
znf296
|
zinc finger protein 296 |
| chr5_-_32396929 | 0.31 |
ENSDART00000023977
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr15_-_39969988 | 0.31 |
ENSDART00000146054
|
rps5
|
ribosomal protein S5 |
| chr7_+_66634167 | 0.30 |
ENSDART00000027616
|
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr2_+_1988036 | 0.30 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr15_-_8517376 | 0.30 |
ENSDART00000186289
|
npas1
|
neuronal PAS domain protein 1 |
| chr6_-_10728921 | 0.30 |
ENSDART00000151484
|
sp3b
|
Sp3b transcription factor |
| chr19_+_28187480 | 0.30 |
ENSDART00000183825
|
irx4b
|
iroquois homeobox 4b |
| chr6_+_23057311 | 0.29 |
ENSDART00000026448
|
evpla
|
envoplakin a |
| chr4_+_53976731 | 0.29 |
ENSDART00000165813
|
si:ch211-249c2.1
|
si:ch211-249c2.1 |
| chr9_+_21795917 | 0.29 |
ENSDART00000169069
|
rev1
|
REV1, polymerase (DNA directed) |
| chr14_+_38656776 | 0.29 |
ENSDART00000186197
ENSDART00000185861 ENSDART00000020625 |
fbxo38
|
F-box protein 38 |
| chr6_-_10728057 | 0.29 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr2_-_42628028 | 0.28 |
ENSDART00000179866
|
myo10
|
myosin X |
| chr11_-_35171162 | 0.28 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr16_+_26706519 | 0.28 |
ENSDART00000142706
|
virma
|
vir like m6A methyltransferase associated |
| chr20_-_21672970 | 0.28 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr5_-_51998708 | 0.28 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr18_-_10995410 | 0.27 |
ENSDART00000136751
|
tspan33b
|
tetraspanin 33b |
| chr5_-_38451082 | 0.27 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr5_+_25762271 | 0.27 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr12_+_20693743 | 0.27 |
ENSDART00000153023
ENSDART00000153370 |
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr19_+_42770041 | 0.27 |
ENSDART00000150930
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr5_+_32835219 | 0.27 |
ENSDART00000140832
ENSDART00000186055 |
si:ch211-208h16.4
|
si:ch211-208h16.4 |
| chr7_-_48263516 | 0.27 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr9_+_38168012 | 0.26 |
ENSDART00000102445
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr8_-_44298964 | 0.26 |
ENSDART00000098520
|
fzd10
|
frizzled class receptor 10 |
| chr23_-_16692312 | 0.26 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr7_-_50367642 | 0.26 |
ENSDART00000134941
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr21_+_5974590 | 0.26 |
ENSDART00000098499
|
dck
|
deoxycytidine kinase |
| chr14_-_16754262 | 0.26 |
ENSDART00000001159
|
mgat4b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr15_-_18176694 | 0.26 |
ENSDART00000189840
|
tmprss5
|
transmembrane protease, serine 5 |
| chr23_+_28494189 | 0.26 |
ENSDART00000146990
ENSDART00000006657 |
huwe1
|
HECT, UBA and WWE domain containing 1 |
| chr5_-_40734045 | 0.26 |
ENSDART00000010896
|
isl1
|
ISL LIM homeobox 1 |
| chr11_+_24314148 | 0.26 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr2_+_36620011 | 0.26 |
ENSDART00000177428
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr13_+_11876437 | 0.26 |
ENSDART00000179753
|
trim8a
|
tripartite motif containing 8a |
| chr23_-_33738570 | 0.25 |
ENSDART00000131680
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr24_-_9300160 | 0.25 |
ENSDART00000152378
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr7_+_37742299 | 0.25 |
ENSDART00000143300
|
brd7
|
bromodomain containing 7 |
| chr22_-_22231720 | 0.25 |
ENSDART00000160165
|
ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr19_+_29808699 | 0.25 |
ENSDART00000051799
ENSDART00000164205 |
hdac1
|
histone deacetylase 1 |
| chr8_+_29742237 | 0.25 |
ENSDART00000133955
ENSDART00000020621 |
mapk4
|
mitogen-activated protein kinase 4 |
| chr23_-_18057851 | 0.24 |
ENSDART00000173075
ENSDART00000173230 ENSDART00000173135 ENSDART00000173431 ENSDART00000173068 ENSDART00000172987 |
zgc:92287
|
zgc:92287 |
| chr17_+_8799661 | 0.24 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr14_+_6423973 | 0.24 |
ENSDART00000051556
|
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr9_-_28939181 | 0.24 |
ENSDART00000101276
ENSDART00000135334 |
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr24_+_24461558 | 0.24 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr12_-_48168135 | 0.24 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr6_+_2097690 | 0.24 |
ENSDART00000193770
|
tgm2b
|
transglutaminase 2b |
| chr10_+_25355308 | 0.24 |
ENSDART00000100415
|
map3k7cl
|
map3k7 C-terminal like |
| chr11_-_44999858 | 0.24 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr1_+_35473219 | 0.23 |
ENSDART00000109678
ENSDART00000181635 |
usp38
|
ubiquitin specific peptidase 38 |
| chr1_-_29747702 | 0.23 |
ENSDART00000133225
ENSDART00000189670 |
spp2
|
secreted phosphoprotein 2 |
| chr17_+_8799451 | 0.23 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr16_+_28764017 | 0.23 |
ENSDART00000122433
|
trim33l
|
tripartite motif containing 33, like |
| chr24_+_26276805 | 0.23 |
ENSDART00000089749
|
adipoqa
|
adiponectin, C1Q and collagen domain containing, a |
| chr19_+_29808471 | 0.23 |
ENSDART00000186428
|
hdac1
|
histone deacetylase 1 |
| chr16_-_26676685 | 0.23 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr23_-_18057270 | 0.22 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr19_-_32150078 | 0.22 |
ENSDART00000134934
ENSDART00000186410 ENSDART00000181780 |
pag1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr16_+_28728347 | 0.22 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr8_+_14381272 | 0.22 |
ENSDART00000057642
|
acbd6
|
acyl-CoA binding domain containing 6 |
| chr16_+_24721556 | 0.22 |
ENSDART00000155754
|
smg9
|
smg9 nonsense mediated mRNA decay factor |
| chr17_-_21066075 | 0.22 |
ENSDART00000078763
ENSDART00000104327 |
vsx1
|
visual system homeobox 1 homolog, chx10-like |
| chr20_+_43691208 | 0.22 |
ENSDART00000152976
ENSDART00000045185 |
lin9
|
lin-9 DREAM MuvB core complex component |
| chr5_-_22019061 | 0.22 |
ENSDART00000113066
|
amer1
|
APC membrane recruitment protein 1 |
| chr18_-_19819812 | 0.21 |
ENSDART00000060344
|
aagab
|
alpha and gamma adaptin binding protein |
| chr13_+_24287093 | 0.21 |
ENSDART00000058628
|
ccsapb
|
centriole, cilia and spindle-associated protein b |
| chr13_+_11876145 | 0.21 |
ENSDART00000128249
|
trim8a
|
tripartite motif containing 8a |
| chr10_+_15777258 | 0.21 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr3_-_28075756 | 0.21 |
ENSDART00000122037
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr2_-_15040345 | 0.21 |
ENSDART00000109657
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr18_+_30441740 | 0.21 |
ENSDART00000189074
|
gse1
|
Gse1 coiled-coil protein |
| chr16_-_13623659 | 0.20 |
ENSDART00000168978
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr11_+_24313931 | 0.20 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr2_+_32016516 | 0.20 |
ENSDART00000135040
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr7_-_18470963 | 0.20 |
ENSDART00000173929
ENSDART00000173638 |
znf16l
|
zinc finger protein 16 like |
| chr7_+_20471315 | 0.20 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr7_+_6969909 | 0.20 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr20_-_37831849 | 0.20 |
ENSDART00000188483
ENSDART00000153005 ENSDART00000142364 |
si:ch211-147d7.5
|
si:ch211-147d7.5 |
| chr24_+_20575259 | 0.19 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr23_-_19484160 | 0.19 |
ENSDART00000137026
|
fam208ab
|
family with sequence similarity 208, member Ab |
| chr7_-_69795488 | 0.19 |
ENSDART00000162414
|
USP53 (1 of many)
|
ubiquitin specific peptidase 53 |
| chr5_-_30475011 | 0.19 |
ENSDART00000187501
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr10_+_29850330 | 0.19 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr7_+_31879649 | 0.19 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr16_+_30933044 | 0.18 |
ENSDART00000184453
|
gstk2
|
glutathione S-transferase kappa 2 |
| chr7_-_41726657 | 0.18 |
ENSDART00000099121
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr10_+_2841205 | 0.18 |
ENSDART00000131505
ENSDART00000055869 |
ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr7_-_68373495 | 0.18 |
ENSDART00000167440
|
zfhx3
|
zinc finger homeobox 3 |
| chr1_-_17650223 | 0.18 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr13_+_2894536 | 0.18 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr4_-_23858900 | 0.17 |
ENSDART00000123199
|
usp6nl
|
USP6 N-terminal like |
| chr6_-_2454691 | 0.17 |
ENSDART00000192820
|
MED8
|
mediator complex subunit 8 |
| chr1_+_27825980 | 0.17 |
ENSDART00000160524
|
pspc1
|
paraspeckle component 1 |
| chr5_+_57924611 | 0.17 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr25_+_36674715 | 0.17 |
ENSDART00000111861
|
mafb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog b (paralog b) |
| chr5_-_13315726 | 0.17 |
ENSDART00000143364
|
sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr3_-_49514874 | 0.17 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr11_-_36450770 | 0.17 |
ENSDART00000128889
|
zbtb40
|
zinc finger and BTB domain containing 40 |
| chr11_-_41966854 | 0.17 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr19_-_20446756 | 0.17 |
ENSDART00000140711
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr10_-_6867282 | 0.16 |
ENSDART00000144001
ENSDART00000109744 |
ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr20_-_39391833 | 0.16 |
ENSDART00000135149
|
si:dkey-217m5.8
|
si:dkey-217m5.8 |
| chr18_-_26797723 | 0.16 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr11_+_18873113 | 0.16 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr9_-_98982 | 0.16 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr16_-_13623928 | 0.16 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr4_+_12615836 | 0.16 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr9_-_8454060 | 0.16 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr5_+_34622320 | 0.15 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr1_-_39895859 | 0.15 |
ENSDART00000135791
ENSDART00000035739 |
tmem134
|
transmembrane protein 134 |
| chr9_-_27717006 | 0.15 |
ENSDART00000146860
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr25_-_7650335 | 0.15 |
ENSDART00000089034
|
myo5ab
|
myosin VAb |
| chr15_+_1397811 | 0.15 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr19_+_40145400 | 0.15 |
ENSDART00000049968
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr19_+_7636941 | 0.15 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr7_+_24251367 | 0.15 |
ENSDART00000173642
|
svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr1_-_26293203 | 0.15 |
ENSDART00000180140
|
cxxc4
|
CXXC finger 4 |
| chr18_+_44566863 | 0.15 |
ENSDART00000176084
|
dhx34
|
DEAH (Asp-Glu-Ala-His) box polypeptide 34 |
| chr22_+_16308806 | 0.14 |
ENSDART00000162685
|
lrrc39
|
leucine rich repeat containing 39 |
| chr2_-_39015130 | 0.14 |
ENSDART00000044331
|
copb2
|
coatomer protein complex, subunit beta 2 |
| chr9_+_30633184 | 0.14 |
ENSDART00000191310
|
tbc1d4
|
TBC1 domain family, member 4 |
| chr19_+_35002479 | 0.14 |
ENSDART00000103266
|
wdyhv1
|
WDYHV motif containing 1 |
| chr16_-_19568795 | 0.14 |
ENSDART00000185141
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr15_-_16177603 | 0.14 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr10_+_29849977 | 0.14 |
ENSDART00000180242
|
hspa8
|
heat shock protein 8 |
| chr25_-_25619550 | 0.14 |
ENSDART00000150400
|
tbc1d2b
|
TBC1 domain family, member 2B |
| chr2_-_42128714 | 0.14 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr10_+_15777064 | 0.14 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr13_-_7575216 | 0.14 |
ENSDART00000159443
|
pitx3
|
paired-like homeodomain 3 |
| chr13_-_14926318 | 0.13 |
ENSDART00000142785
|
cdc25b
|
cell division cycle 25B |
| chr1_+_20593653 | 0.13 |
ENSDART00000132440
|
si:ch211-142c4.1
|
si:ch211-142c4.1 |
| chr9_+_2020667 | 0.13 |
ENSDART00000157818
|
lnpa
|
limb and neural patterns a |
| chr21_+_27382893 | 0.13 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr4_+_8532580 | 0.13 |
ENSDART00000162065
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr1_-_26292897 | 0.13 |
ENSDART00000112899
ENSDART00000185410 |
cxxc4
|
CXXC finger 4 |
| chr23_-_7826849 | 0.12 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr23_-_19831739 | 0.12 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr19_-_33087246 | 0.12 |
ENSDART00000052078
|
klhl38a
|
kelch-like family member 38a |
| chr3_+_20156956 | 0.12 |
ENSDART00000125281
|
ngfra
|
nerve growth factor receptor a (TNFR superfamily, member 16) |
| chr22_+_2260497 | 0.12 |
ENSDART00000142890
|
si:dkey-4c15.9
|
si:dkey-4c15.9 |
| chr11_-_28050559 | 0.12 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr19_+_9666961 | 0.12 |
ENSDART00000104596
ENSDART00000132124 |
si:dkey-14o18.6
|
si:dkey-14o18.6 |
| chr7_+_7048245 | 0.12 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr21_-_45363871 | 0.11 |
ENSDART00000075443
ENSDART00000182078 ENSDART00000151106 |
PPP2CA
|
zgc:56064 |
| chr12_-_31103187 | 0.11 |
ENSDART00000005562
ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 ENSDART00000122972 ENSDART00000153068 |
tcf7l2
|
transcription factor 7 like 2 |
| chr13_+_1575276 | 0.11 |
ENSDART00000165987
|
DST
|
dystonin |
| chr3_-_15889508 | 0.11 |
ENSDART00000148363
|
cramp1
|
cramped chromatin regulator homolog 1 |
| chr14_-_15699528 | 0.11 |
ENSDART00000161123
|
neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr20_+_20902549 | 0.11 |
ENSDART00000181195
|
brf1b
|
BRF1, RNA polymerase III transcription initiation factor b |
| chr3_+_23654233 | 0.11 |
ENSDART00000078428
|
hoxb13a
|
homeobox B13a |
| chr18_+_19820008 | 0.11 |
ENSDART00000079691
ENSDART00000135049 |
iqch
|
IQ motif containing H |
| chr20_-_22798794 | 0.11 |
ENSDART00000148084
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of msc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 1.3 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 0.5 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.6 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.4 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.3 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.2 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.1 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.5 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.1 | 0.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.3 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.1 | 0.3 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.0 | 0.1 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.2 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.1 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 0.2 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.4 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.0 | 0.2 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0021744 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.0 | 0.1 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.3 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.1 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.1 | GO:1900136 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.1 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.4 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.1 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.3 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.4 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.3 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.2 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.3 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.3 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.6 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0071712 | protein O-linked mannosylation(GO:0035269) ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.3 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.2 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.1 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 1.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.4 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.3 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.2 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0042164 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |