Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
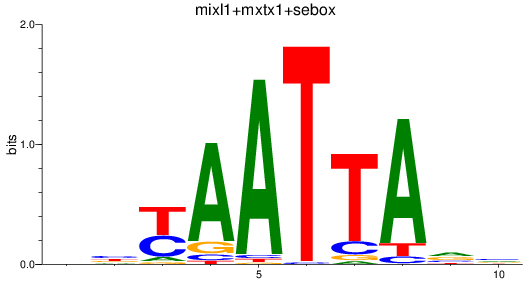
Results for mixl1+mxtx1+sebox
Z-value: 0.39
Transcription factors associated with mixl1+mxtx1+sebox
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sebox
|
ENSDARG00000042526 | SEBOX homeobox |
|
mixl1
|
ENSDARG00000069252 | Mix paired-like homeobox |
|
mxtx1
|
ENSDARG00000069382 | mix-type homeobox gene 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mxtx1 | dr11_v1_chr13_-_21660203_21660203 | 0.92 | 3.4e-08 | Click! |
| sebox | dr11_v1_chr5_+_67371650_67371650 | -0.49 | 3.3e-02 | Click! |
| mixl1 | dr11_v1_chr20_-_43723860_43723860 | 0.44 | 6.0e-02 | Click! |
Activity profile of mixl1+mxtx1+sebox motif
Sorted Z-values of mixl1+mxtx1+sebox motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_15440841 | 1.26 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr16_+_29509133 | 1.25 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr19_+_15441022 | 1.06 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr23_-_17003533 | 0.98 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr1_-_55248496 | 0.94 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr21_-_23331619 | 0.81 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr17_+_16046132 | 0.76 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr17_+_16046314 | 0.73 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr18_+_19456648 | 0.71 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr5_-_12219572 | 0.66 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr17_+_21295132 | 0.65 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr11_-_35171162 | 0.60 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr2_+_33326522 | 0.58 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr14_+_22113331 | 0.58 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr16_-_29387215 | 0.58 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr17_-_16422654 | 0.58 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr6_+_41191482 | 0.58 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr16_-_16761164 | 0.56 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr23_+_31912882 | 0.55 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr6_-_40922971 | 0.54 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr15_+_36309070 | 0.54 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr22_+_16535575 | 0.52 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr12_-_35830625 | 0.52 |
ENSDART00000180028
|
CU459056.1
|
|
| chr8_+_23355484 | 0.51 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr3_+_18807006 | 0.50 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr14_-_22113600 | 0.50 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr7_+_56098590 | 0.49 |
ENSDART00000098453
|
cdh15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr22_-_15593824 | 0.49 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr13_+_35637048 | 0.48 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr10_-_21362320 | 0.48 |
ENSDART00000189789
|
avd
|
avidin |
| chr21_+_43404945 | 0.47 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr16_-_17347727 | 0.46 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr17_+_8799451 | 0.46 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr10_-_21362071 | 0.45 |
ENSDART00000125167
|
avd
|
avidin |
| chr24_+_19415124 | 0.45 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr1_+_21731382 | 0.45 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr21_+_15592426 | 0.45 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr23_-_31913069 | 0.45 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr20_-_23426339 | 0.44 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr13_+_27232848 | 0.44 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr8_-_37249813 | 0.44 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr23_+_31913292 | 0.43 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr2_+_6253246 | 0.43 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr2_+_20793982 | 0.43 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr21_+_34088110 | 0.42 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr2_+_41526904 | 0.41 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr23_-_31913231 | 0.40 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr4_+_11723852 | 0.40 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr20_-_20930926 | 0.40 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr6_+_40922572 | 0.40 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr3_-_16719244 | 0.39 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr17_-_31695217 | 0.39 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr14_-_33945692 | 0.38 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr3_+_28860283 | 0.38 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr13_+_22295905 | 0.38 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr13_-_31017960 | 0.38 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr2_-_55298075 | 0.37 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr11_+_33818179 | 0.37 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr3_+_26244353 | 0.37 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr11_-_30158191 | 0.36 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr4_+_9177997 | 0.36 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr24_+_21540842 | 0.36 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr19_+_30884706 | 0.35 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr16_+_28994709 | 0.35 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr19_+_2631565 | 0.34 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr15_+_1534644 | 0.33 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr14_+_6962271 | 0.33 |
ENSDART00000148447
ENSDART00000149114 ENSDART00000149492 ENSDART00000148394 |
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr9_+_29548195 | 0.32 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr15_+_857148 | 0.32 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr6_-_8311044 | 0.32 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr15_-_2519640 | 0.32 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr11_-_41220794 | 0.31 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr1_-_45616470 | 0.31 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr3_-_30488063 | 0.31 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr24_-_4450238 | 0.31 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr3_-_39695856 | 0.31 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr21_-_17296789 | 0.31 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr20_-_20931197 | 0.31 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr23_+_11285662 | 0.31 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr23_-_25135046 | 0.31 |
ENSDART00000184844
ENSDART00000103989 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr5_+_35786141 | 0.30 |
ENSDART00000022043
ENSDART00000127383 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr8_+_50953776 | 0.29 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr13_+_25486608 | 0.29 |
ENSDART00000057689
|
bag3
|
BCL2 associated athanogene 3 |
| chr16_-_42056137 | 0.29 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr24_-_2450597 | 0.29 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr2_-_30668580 | 0.29 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr6_+_21001264 | 0.29 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr3_-_26244256 | 0.27 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr16_+_42471455 | 0.27 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr11_+_35171406 | 0.27 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr5_+_52167986 | 0.26 |
ENSDART00000162256
ENSDART00000073626 |
slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr23_+_39695827 | 0.26 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr24_-_37640705 | 0.26 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr4_-_5455506 | 0.26 |
ENSDART00000156593
ENSDART00000154676 |
si:dkey-14d8.22
|
si:dkey-14d8.22 |
| chr14_-_33481428 | 0.26 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr10_+_6884627 | 0.26 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr9_+_25776971 | 0.25 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr13_-_12602920 | 0.25 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr11_-_45138857 | 0.25 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr5_-_14326959 | 0.25 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr15_-_2493771 | 0.25 |
ENSDART00000184906
|
neu4
|
sialidase 4 |
| chr11_+_31864921 | 0.24 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr17_-_40956035 | 0.24 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr1_-_26444075 | 0.24 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr1_-_43727012 | 0.24 |
ENSDART00000181064
|
bdh2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr9_-_31278048 | 0.24 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr14_+_34490445 | 0.24 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr21_+_25777425 | 0.24 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr13_+_1132261 | 0.24 |
ENSDART00000146049
|
wdr92
|
WD repeat domain 92 |
| chr12_+_16087077 | 0.23 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr15_-_18115540 | 0.23 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr16_+_33902006 | 0.23 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr12_+_48803098 | 0.23 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr21_-_11654422 | 0.23 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr16_-_43344859 | 0.23 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr17_-_25649079 | 0.23 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr3_-_39696066 | 0.23 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr24_+_32668675 | 0.22 |
ENSDART00000156638
ENSDART00000155973 |
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr3_+_46635527 | 0.22 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr11_-_30634286 | 0.22 |
ENSDART00000191019
|
zgc:153665
|
zgc:153665 |
| chr17_-_20118145 | 0.22 |
ENSDART00000149737
ENSDART00000165606 |
ryr2b
|
ryanodine receptor 2b (cardiac) |
| chr3_+_45365098 | 0.22 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr15_-_16177603 | 0.21 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr13_-_36535128 | 0.21 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr6_-_12172424 | 0.20 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr18_+_30028135 | 0.20 |
ENSDART00000140825
ENSDART00000145306 ENSDART00000136810 |
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr8_+_45334255 | 0.19 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr17_+_46818521 | 0.19 |
ENSDART00000156022
|
pimr14
|
Pim proto-oncogene, serine/threonine kinase, related 14 |
| chr3_+_45364849 | 0.19 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr8_+_41037541 | 0.19 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr7_+_34592526 | 0.19 |
ENSDART00000173959
|
fhod1
|
formin homology 2 domain containing 1 |
| chr4_-_20292821 | 0.19 |
ENSDART00000136069
ENSDART00000192504 |
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr8_-_37249991 | 0.19 |
ENSDART00000189275
ENSDART00000178556 |
rbm39b
|
RNA binding motif protein 39b |
| chr1_+_33668236 | 0.18 |
ENSDART00000122316
ENSDART00000102184 |
arl13b
|
ADP-ribosylation factor-like 13b |
| chr5_+_28770273 | 0.18 |
ENSDART00000114473
|
trafd1
|
TRAF-type zinc finger domain containing 1 |
| chr7_+_19495379 | 0.17 |
ENSDART00000180514
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr18_-_19456269 | 0.17 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr21_-_36453594 | 0.17 |
ENSDART00000193176
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr11_+_26604224 | 0.17 |
ENSDART00000030453
ENSDART00000168895 ENSDART00000159505 |
dynlrb1
|
dynein, light chain, roadblock-type 1 |
| chr19_+_40069524 | 0.17 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr19_-_30524952 | 0.17 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr9_-_20372977 | 0.16 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr17_-_23416897 | 0.16 |
ENSDART00000163391
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr15_+_34988148 | 0.16 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr8_+_21353878 | 0.16 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr21_-_25801956 | 0.15 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr12_-_14143344 | 0.15 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr16_+_6750756 | 0.15 |
ENSDART00000149720
|
znf236
|
zinc finger protein 236 |
| chr24_-_30862168 | 0.15 |
ENSDART00000168540
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr2_-_57378748 | 0.15 |
ENSDART00000149235
|
tcf3a
|
transcription factor 3a |
| chr3_-_30685401 | 0.14 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr24_+_12835935 | 0.14 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr4_-_43388943 | 0.14 |
ENSDART00000150796
|
si:dkey-29j8.2
|
si:dkey-29j8.2 |
| chr15_-_43284021 | 0.14 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr25_-_2723682 | 0.13 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr21_-_35419486 | 0.13 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr8_-_25033681 | 0.13 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr24_+_9412450 | 0.13 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr21_+_34088377 | 0.12 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr7_+_26649319 | 0.12 |
ENSDART00000173823
ENSDART00000101053 |
tp53i11a
|
tumor protein p53 inducible protein 11a |
| chr9_-_21918963 | 0.12 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr6_+_50381665 | 0.12 |
ENSDART00000141128
|
cyc1
|
cytochrome c-1 |
| chr19_-_19379084 | 0.12 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr11_+_18873113 | 0.11 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr3_+_32365811 | 0.11 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr18_-_29896367 | 0.11 |
ENSDART00000191303
|
cmc2
|
C-x(9)-C motif containing 2 |
| chr5_-_20491198 | 0.11 |
ENSDART00000183051
ENSDART00000144232 |
ficd
|
FIC domain containing |
| chr1_+_8694196 | 0.11 |
ENSDART00000025604
|
zgc:77849
|
zgc:77849 |
| chr10_+_40660772 | 0.11 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr14_+_44794936 | 0.11 |
ENSDART00000128881
|
zgc:195212
|
zgc:195212 |
| chr8_+_25034544 | 0.11 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr14_+_45406299 | 0.10 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr10_+_21867307 | 0.10 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr15_-_41332865 | 0.10 |
ENSDART00000170014
|
si:dkey-121n8.7
|
si:dkey-121n8.7 |
| chr22_+_31207799 | 0.09 |
ENSDART00000133267
|
grip2b
|
glutamate receptor interacting protein 2b |
| chr17_+_37227936 | 0.09 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr5_-_37871526 | 0.09 |
ENSDART00000136450
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr20_+_52554352 | 0.09 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr14_+_34492288 | 0.08 |
ENSDART00000144301
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr6_+_50381347 | 0.08 |
ENSDART00000055504
|
cyc1
|
cytochrome c-1 |
| chr8_+_11425048 | 0.08 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr15_+_5360407 | 0.08 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr18_-_40708537 | 0.08 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr9_-_746317 | 0.07 |
ENSDART00000129632
ENSDART00000130720 |
usp37
|
ubiquitin specific peptidase 37 |
| chr24_-_3783497 | 0.07 |
ENSDART00000158354
|
adarb2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr20_-_9095105 | 0.07 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr4_+_2655358 | 0.07 |
ENSDART00000007638
|
bcap29
|
B cell receptor associated protein 29 |
| chr18_+_24921587 | 0.07 |
ENSDART00000191345
|
rgma
|
repulsive guidance molecule family member a |
| chr24_-_6078222 | 0.07 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr16_-_29458806 | 0.06 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr21_+_5272059 | 0.06 |
ENSDART00000141981
|
loxhd1a
|
lipoxygenase homology domains 1a |
| chr11_-_1550709 | 0.06 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr13_-_24311628 | 0.06 |
ENSDART00000004420
|
rab4a
|
RAB4a, member RAS oncogene family |
| chr17_+_23298928 | 0.06 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr24_-_40860603 | 0.06 |
ENSDART00000188032
|
CU633479.7
|
|
| chr10_-_25217347 | 0.06 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chrM_+_3803 | 0.05 |
ENSDART00000093596
|
mt-nd1
|
NADH dehydrogenase 1, mitochondrial |
| chr21_-_39177564 | 0.05 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr3_+_18398876 | 0.05 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr2_-_32384683 | 0.05 |
ENSDART00000182942
ENSDART00000141757 |
ubtfl
|
upstream binding transcription factor, like |
Network of associatons between targets according to the STRING database.
First level regulatory network of mixl1+mxtx1+sebox
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 0.7 | GO:0098924 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.2 | 1.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.2 | 0.6 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.2 | 0.5 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.6 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.3 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.5 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.3 | GO:0060898 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.4 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 0.4 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.3 | GO:0010660 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.1 | 0.4 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.7 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.7 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 0.8 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.2 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.5 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.3 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.4 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.7 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.3 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.2 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.3 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.3 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.5 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.2 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.4 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.3 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.3 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.3 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 0.3 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.4 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.3 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.0 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.2 | 0.9 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 1.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.4 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.7 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.5 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.6 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.6 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |