Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for meox2a
Z-value: 0.80
Transcription factors associated with meox2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meox2a
|
ENSDARG00000040911 | mesenchyme homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meox2a | dr11_v1_chr15_-_34458495_34458495 | 0.36 | 1.3e-01 | Click! |
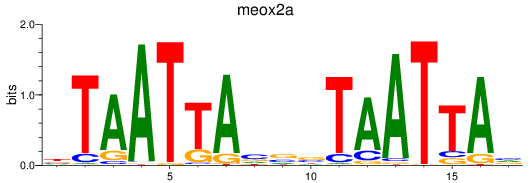
Activity profile of meox2a motif
Sorted Z-values of meox2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_35075847 | 2.20 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr5_-_26118855 | 1.99 |
ENSDART00000009028
|
ela3l
|
elastase 3 like |
| chr2_+_50862527 | 1.66 |
ENSDART00000169800
ENSDART00000158847 ENSDART00000160900 |
adcyap1r1a
|
adenylate cyclase activating polypeptide 1a (pituitary) receptor type I |
| chr5_+_2815021 | 1.31 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr1_-_43905252 | 1.22 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr11_+_43043171 | 1.18 |
ENSDART00000180344
|
CABZ01092982.1
|
|
| chr18_+_7286788 | 1.05 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr6_-_55585423 | 1.04 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr23_-_306796 | 0.97 |
ENSDART00000143125
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr16_+_20161805 | 0.93 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr4_-_77506362 | 0.91 |
ENSDART00000174387
ENSDART00000181181 |
CABZ01087415.1
|
|
| chr3_+_39540014 | 0.89 |
ENSDART00000074848
|
zgc:165423
|
zgc:165423 |
| chr15_+_45563656 | 0.89 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr19_-_29887629 | 0.83 |
ENSDART00000066123
|
kpna6
|
karyopherin alpha 6 (importin alpha 7) |
| chr8_+_39724138 | 0.82 |
ENSDART00000009323
|
pla2g1b
|
phospholipase A2, group IB (pancreas) |
| chr23_-_3511630 | 0.80 |
ENSDART00000019667
|
rnf114
|
ring finger protein 114 |
| chr17_-_47090440 | 0.79 |
ENSDART00000163542
|
CABZ01056321.1
|
|
| chr22_-_21845685 | 0.74 |
ENSDART00000105564
|
aes
|
amino-terminal enhancer of split |
| chr4_+_5531583 | 0.73 |
ENSDART00000137500
ENSDART00000042080 |
si:dkey-14d8.6
|
si:dkey-14d8.6 |
| chr7_-_71758307 | 0.72 |
ENSDART00000161067
ENSDART00000165253 |
myom1b
|
myomesin 1b |
| chr16_-_17175731 | 0.72 |
ENSDART00000183057
|
opn9
|
opsin 9 |
| chr19_-_40192249 | 0.72 |
ENSDART00000051972
|
grn1
|
granulin 1 |
| chrM_+_11009 | 0.71 |
ENSDART00000093617
|
mt-nd4l
|
NADH dehydrogenase 4L, mitochondrial |
| chr24_-_29963858 | 0.69 |
ENSDART00000183442
|
CR352310.1
|
|
| chr16_+_14588141 | 0.69 |
ENSDART00000140469
ENSDART00000059984 ENSDART00000167411 ENSDART00000133566 |
deptor
|
DEP domain containing MTOR-interacting protein |
| chr13_+_28701233 | 0.69 |
ENSDART00000135931
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr8_-_40555340 | 0.67 |
ENSDART00000163348
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr15_+_6114109 | 0.65 |
ENSDART00000184937
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr18_-_341489 | 0.64 |
ENSDART00000168333
|
si:ch1073-83b15.1
|
si:ch1073-83b15.1 |
| chr20_-_40755614 | 0.63 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr15_+_9053059 | 0.62 |
ENSDART00000012039
|
ppm1na
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Na (putative) |
| chr10_-_44508249 | 0.61 |
ENSDART00000160018
|
DUSP26
|
dual specificity phosphatase 26 |
| chr2_+_56937548 | 0.61 |
ENSDART00000189308
|
CABZ01117752.1
|
|
| chr19_+_9295244 | 0.61 |
ENSDART00000132255
ENSDART00000144299 |
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr3_-_32169754 | 0.60 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr19_+_823945 | 0.60 |
ENSDART00000142287
|
ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr18_+_11506561 | 0.59 |
ENSDART00000121647
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr4_-_17793152 | 0.57 |
ENSDART00000134080
|
mybpc1
|
myosin binding protein C, slow type |
| chr4_-_277081 | 0.56 |
ENSDART00000166174
|
si:ch73-252i11.1
|
si:ch73-252i11.1 |
| chr2_+_33382648 | 0.56 |
ENSDART00000137207
ENSDART00000098831 |
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr14_+_33413980 | 0.56 |
ENSDART00000052780
ENSDART00000124437 ENSDART00000173327 |
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr11_+_22374419 | 0.54 |
ENSDART00000174683
ENSDART00000170521 ENSDART00000193980 |
ppfia4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr21_+_8427059 | 0.54 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr23_+_14696043 | 0.54 |
ENSDART00000132037
|
si:rp71-79p20.2
|
si:rp71-79p20.2 |
| chr19_+_31873308 | 0.54 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr25_+_31267268 | 0.53 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr7_-_72208248 | 0.53 |
ENSDART00000108916
|
zmp:0000001168
|
zmp:0000001168 |
| chr9_-_42418470 | 0.53 |
ENSDART00000144353
|
calcrla
|
calcitonin receptor-like a |
| chr2_+_57801960 | 0.51 |
ENSDART00000147966
|
si:dkeyp-68b7.10
|
si:dkeyp-68b7.10 |
| chr15_+_45563491 | 0.51 |
ENSDART00000191169
|
cldn15lb
|
claudin 15-like b |
| chr9_-_19699728 | 0.51 |
ENSDART00000166780
|
si:ch211-141e20.2
|
si:ch211-141e20.2 |
| chr8_-_13972626 | 0.51 |
ENSDART00000144296
|
serping1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr3_-_25268751 | 0.50 |
ENSDART00000139423
|
mgat3a
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase a |
| chr22_+_1539373 | 0.50 |
ENSDART00000172104
|
si:ch211-255f4.7
|
si:ch211-255f4.7 |
| chr9_-_1703761 | 0.50 |
ENSDART00000144822
ENSDART00000137210 ENSDART00000135273 |
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr18_+_9615147 | 0.48 |
ENSDART00000160284
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr21_-_5958387 | 0.48 |
ENSDART00000130521
|
CACFD1
|
si:ch73-209e20.5 |
| chr9_+_46644633 | 0.48 |
ENSDART00000160285
|
slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr20_+_1960092 | 0.47 |
ENSDART00000191892
|
CABZ01103860.1
|
|
| chr10_-_35236949 | 0.47 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr13_-_23264724 | 0.47 |
ENSDART00000051886
|
si:dkey-103j14.5
|
si:dkey-103j14.5 |
| chr3_+_26019426 | 0.46 |
ENSDART00000135389
ENSDART00000182411 |
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr11_+_2506516 | 0.46 |
ENSDART00000130886
ENSDART00000189767 |
NABP2
|
si:ch73-190f16.2 |
| chr5_-_44286987 | 0.46 |
ENSDART00000184112
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr5_+_69981453 | 0.46 |
ENSDART00000143860
|
si:ch211-154e10.1
|
si:ch211-154e10.1 |
| chr1_-_22512063 | 0.45 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr17_-_4252221 | 0.44 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr11_-_11266882 | 0.44 |
ENSDART00000020256
|
lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr5_+_23622177 | 0.43 |
ENSDART00000121504
|
cx27.5
|
connexin 27.5 |
| chr11_-_1935883 | 0.43 |
ENSDART00000172885
|
faim2b
|
Fas apoptotic inhibitory molecule 2b |
| chr19_+_5072918 | 0.43 |
ENSDART00000037126
|
eno2
|
enolase 2 |
| chr5_-_37117778 | 0.43 |
ENSDART00000149138
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr23_+_45538932 | 0.43 |
ENSDART00000135602
|
si:ch73-290k24.6
|
si:ch73-290k24.6 |
| chr16_-_24642814 | 0.42 |
ENSDART00000153987
ENSDART00000154319 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr25_+_2361721 | 0.41 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr21_+_26071874 | 0.41 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr12_-_3753131 | 0.41 |
ENSDART00000129668
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr5_+_20035284 | 0.41 |
ENSDART00000191808
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr3_+_28953274 | 0.41 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr9_-_8661436 | 0.41 |
ENSDART00000130442
|
col4a1
|
collagen, type IV, alpha 1 |
| chr9_-_51323545 | 0.40 |
ENSDART00000139316
|
slc4a10b
|
solute carrier family 4, sodium bicarbonate transporter, member 10b |
| chr21_-_43474012 | 0.40 |
ENSDART00000065104
|
tmem185
|
transmembrane protein 185 |
| chr24_-_33276139 | 0.39 |
ENSDART00000128943
|
nrbp2b
|
nuclear receptor binding protein 2b |
| chr11_-_2131280 | 0.39 |
ENSDART00000008409
|
calcoco1b
|
calcium binding and coiled-coil domain 1b |
| chr1_-_9195629 | 0.39 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr16_+_43344475 | 0.39 |
ENSDART00000085282
|
mrpl32
|
mitochondrial ribosomal protein L32 |
| chr25_-_8030113 | 0.38 |
ENSDART00000104674
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr13_-_29505604 | 0.38 |
ENSDART00000110005
|
cdhr1a
|
cadherin-related family member 1a |
| chr7_+_69905307 | 0.38 |
ENSDART00000035907
|
sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr18_-_399554 | 0.38 |
ENSDART00000164374
ENSDART00000186311 ENSDART00000181816 ENSDART00000181892 |
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr2_+_24188502 | 0.38 |
ENSDART00000181955
ENSDART00000191706 ENSDART00000125909 |
map4l
|
microtubule associated protein 4 like |
| chr20_-_1378514 | 0.37 |
ENSDART00000181830
|
scara5
|
scavenger receptor class A, member 5 (putative) |
| chr11_-_33857911 | 0.37 |
ENSDART00000165370
|
nxph2b
|
neurexophilin 2b |
| chr8_-_14121634 | 0.37 |
ENSDART00000184946
|
bgna
|
biglycan a |
| chr24_+_9178064 | 0.37 |
ENSDART00000142971
|
dlgap1b
|
discs, large (Drosophila) homolog-associated protein 1b |
| chr15_-_44077937 | 0.37 |
ENSDART00000110112
|
lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr1_-_45920632 | 0.37 |
ENSDART00000140890
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr24_+_14937205 | 0.37 |
ENSDART00000091735
|
dok6
|
docking protein 6 |
| chr6_+_2195625 | 0.37 |
ENSDART00000155659
|
acvr1bb
|
activin A receptor type 1Bb |
| chr21_-_13123176 | 0.37 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr6_+_41038757 | 0.36 |
ENSDART00000011245
|
entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr3_-_24458281 | 0.36 |
ENSDART00000153993
|
baiap2l2a
|
BAI1-associated protein 2-like 2a |
| chr19_+_3826782 | 0.36 |
ENSDART00000169222
|
oscp1a
|
organic solute carrier partner 1a |
| chr13_-_18122333 | 0.35 |
ENSDART00000128748
|
washc2c
|
WASH complex subunit 2C |
| chr1_+_12177195 | 0.35 |
ENSDART00000146842
ENSDART00000142081 |
stra6l
|
STRA6-like |
| chr15_-_26844591 | 0.35 |
ENSDART00000077582
|
pitpnm3
|
PITPNM family member 3 |
| chr18_+_924949 | 0.35 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr19_-_27858033 | 0.35 |
ENSDART00000103898
ENSDART00000144884 |
srd5a1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) |
| chr24_+_33802528 | 0.35 |
ENSDART00000136040
ENSDART00000147499 ENSDART00000182322 |
atg9b
|
autophagy related 9B |
| chr23_+_16620801 | 0.35 |
ENSDART00000189859
ENSDART00000184578 |
snphb
|
syntaphilin b |
| chr21_-_2232640 | 0.35 |
ENSDART00000157754
|
zgc:113343
|
zgc:113343 |
| chr19_-_28360033 | 0.34 |
ENSDART00000186994
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr17_+_17955063 | 0.34 |
ENSDART00000104999
|
ccdc85ca
|
coiled-coil domain containing 85C, a |
| chr12_+_17504559 | 0.34 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr12_+_18681477 | 0.34 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr2_-_144393 | 0.34 |
ENSDART00000156008
|
adcy1b
|
adenylate cyclase 1b |
| chr21_+_26612777 | 0.34 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr10_-_8053753 | 0.33 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr21_-_5881344 | 0.33 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr11_-_30158191 | 0.33 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr25_-_18140305 | 0.33 |
ENSDART00000180222
|
kitlga
|
kit ligand a |
| chr3_-_13921173 | 0.33 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr5_+_69950882 | 0.33 |
ENSDART00000097359
|
dnajc25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr24_-_7321928 | 0.32 |
ENSDART00000167570
ENSDART00000045150 |
actr3b
|
ARP3 actin related protein 3 homolog B |
| chr9_-_48407408 | 0.32 |
ENSDART00000058248
|
zgc:172182
|
zgc:172182 |
| chr10_+_44699734 | 0.32 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr3_+_431208 | 0.31 |
ENSDART00000154296
ENSDART00000048733 |
si:ch73-308m11.1
si:dkey-167k11.5
|
si:ch73-308m11.1 si:dkey-167k11.5 |
| chr10_-_8053385 | 0.31 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr20_-_47051996 | 0.31 |
ENSDART00000153330
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr12_+_10631266 | 0.31 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr20_-_42102416 | 0.31 |
ENSDART00000186378
ENSDART00000188253 ENSDART00000186458 |
slc35f1
|
solute carrier family 35, member F1 |
| chr23_-_271722 | 0.31 |
ENSDART00000092886
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr21_-_11655010 | 0.30 |
ENSDART00000144370
ENSDART00000139814 ENSDART00000139289 |
cast
|
calpastatin |
| chr10_-_8060573 | 0.30 |
ENSDART00000147104
ENSDART00000099030 |
si:ch211-251f6.6
|
si:ch211-251f6.6 |
| chr21_-_2333357 | 0.29 |
ENSDART00000158743
|
zgc:113348
|
zgc:113348 |
| chr25_+_20077225 | 0.29 |
ENSDART00000136543
|
tnni4b.1
|
troponin I4b, tandem duplicate 1 |
| chr6_+_55032439 | 0.29 |
ENSDART00000164232
ENSDART00000158845 ENSDART00000157584 ENSDART00000026359 ENSDART00000122794 ENSDART00000183742 |
mybphb
|
myosin binding protein Hb |
| chr10_+_44700103 | 0.29 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr5_-_22590124 | 0.29 |
ENSDART00000172353
|
si:dkey-103e21.5
|
si:dkey-103e21.5 |
| chr21_+_17880511 | 0.29 |
ENSDART00000080481
|
rxraa
|
retinoid X receptor, alpha a |
| chr24_-_3783497 | 0.29 |
ENSDART00000158354
|
adarb2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr1_-_21901589 | 0.29 |
ENSDART00000140553
|
frmpd1a
|
FERM and PDZ domain containing 1a |
| chr18_-_2727764 | 0.29 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr13_-_37608441 | 0.28 |
ENSDART00000140230
|
zgc:123068
|
zgc:123068 |
| chr12_-_47580562 | 0.28 |
ENSDART00000153431
|
rgs7b
|
regulator of G protein signaling 7b |
| chr10_+_8885769 | 0.28 |
ENSDART00000139466
|
itga2.2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 2 |
| chr3_-_40068350 | 0.28 |
ENSDART00000185292
|
myo15aa
|
myosin XVAa |
| chr2_+_10709557 | 0.28 |
ENSDART00000183118
ENSDART00000109723 |
evi5a
|
ecotropic viral integration site 5a |
| chr9_-_14683574 | 0.28 |
ENSDART00000144022
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr1_+_31110817 | 0.28 |
ENSDART00000137863
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr10_+_11261576 | 0.27 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr11_+_11030908 | 0.27 |
ENSDART00000111091
|
pla2r1
|
phospholipase A2 receptor 1 |
| chr8_+_36942262 | 0.27 |
ENSDART00000188173
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr1_-_25370281 | 0.27 |
ENSDART00000050445
|
trim2a
|
tripartite motif containing 2a |
| chr7_+_14632157 | 0.27 |
ENSDART00000161264
|
ntrk3b
|
neurotrophic tyrosine kinase, receptor, type 3b |
| chr13_+_15190677 | 0.27 |
ENSDART00000142240
ENSDART00000129045 |
mavs
|
mitochondrial antiviral signaling protein |
| chr10_+_23548674 | 0.27 |
ENSDART00000079686
|
zmp:0000001103
|
zmp:0000001103 |
| chr5_+_20257225 | 0.27 |
ENSDART00000127919
|
ssh1a
|
slingshot protein phosphatase 1a |
| chr17_-_7818944 | 0.27 |
ENSDART00000135538
ENSDART00000037541 |
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr20_-_20182871 | 0.27 |
ENSDART00000103532
|
kcnh5a
|
potassium voltage-gated channel, subfamily H (eag-related), member 5a |
| chr14_+_3287740 | 0.26 |
ENSDART00000186290
|
cdx1a
|
caudal type homeobox 1a |
| chr9_-_54248182 | 0.26 |
ENSDART00000129540
|
larsa
|
leucyl-tRNA synthetase a |
| chr21_+_1381276 | 0.26 |
ENSDART00000192907
|
TCF4
|
transcription factor 4 |
| chr10_+_5203532 | 0.26 |
ENSDART00000165018
|
cdc42se2
|
CDC42 small effector 2 |
| chr7_+_65261576 | 0.26 |
ENSDART00000169566
|
bco1
|
beta-carotene oxygenase 1 |
| chr23_-_32100106 | 0.26 |
ENSDART00000044658
|
letmd1
|
LETM1 domain containing 1 |
| chr7_+_35245607 | 0.25 |
ENSDART00000193422
ENSDART00000173888 |
amfrb
|
autocrine motility factor receptor b |
| chr16_+_40024883 | 0.24 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr5_-_22590739 | 0.24 |
ENSDART00000141555
|
si:dkey-103e21.5
|
si:dkey-103e21.5 |
| chr12_-_46112892 | 0.24 |
ENSDART00000187128
ENSDART00000114268 |
zgc:153932
|
zgc:153932 |
| chr8_+_14886452 | 0.24 |
ENSDART00000146589
|
soat1
|
sterol O-acyltransferase 1 |
| chr23_+_4646194 | 0.24 |
ENSDART00000092344
|
LO017700.1
|
|
| chr16_+_16969060 | 0.24 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr14_+_26229056 | 0.24 |
ENSDART00000179045
|
LO018208.1
|
|
| chr23_-_41965557 | 0.24 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr20_+_4060839 | 0.23 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr3_-_3209432 | 0.23 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr5_+_25952340 | 0.23 |
ENSDART00000147188
|
trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr15_-_23793641 | 0.23 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr23_-_45398622 | 0.23 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr7_+_26762958 | 0.23 |
ENSDART00000167956
ENSDART00000134717 |
tspan18a
|
tetraspanin 18a |
| chr12_-_36740306 | 0.23 |
ENSDART00000153259
|
si:ch211-216b21.2
|
si:ch211-216b21.2 |
| chr10_+_40160502 | 0.23 |
ENSDART00000171879
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr21_-_25685739 | 0.22 |
ENSDART00000129619
ENSDART00000101205 |
phkg1b
|
phosphorylase kinase, gamma 1b (muscle) |
| chr11_-_40172295 | 0.22 |
ENSDART00000187329
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr17_+_6452511 | 0.22 |
ENSDART00000064694
|
TATDN3
|
Danio rerio TatD DNase domain containing 3-like (LOC571912), mRNA. |
| chr9_-_52847881 | 0.22 |
ENSDART00000166407
|
trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr16_+_5184402 | 0.22 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr13_-_50234122 | 0.21 |
ENSDART00000164338
|
eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr23_+_30048849 | 0.21 |
ENSDART00000126027
|
uts2a
|
urotensin 2, alpha |
| chr16_-_51888952 | 0.21 |
ENSDART00000186407
ENSDART00000175435 |
rpl28
|
ribosomal protein L28 |
| chr24_+_31655939 | 0.21 |
ENSDART00000187337
|
CABZ01029422.1
|
|
| chr5_-_9073433 | 0.21 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr25_-_8030425 | 0.21 |
ENSDART00000014964
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr9_+_49712868 | 0.21 |
ENSDART00000192969
ENSDART00000183310 |
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr18_+_50880096 | 0.21 |
ENSDART00000169782
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr1_+_22174251 | 0.20 |
ENSDART00000137429
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr11_+_42474694 | 0.20 |
ENSDART00000056048
ENSDART00000184710 |
si:ch1073-165f9.2
|
si:ch1073-165f9.2 |
| chr10_+_15024772 | 0.20 |
ENSDART00000135667
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr8_-_10932206 | 0.20 |
ENSDART00000124313
|
nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of meox2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.2 | 1.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.6 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.2 | 0.5 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.1 | 0.6 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 0.5 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 0.4 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.4 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.1 | GO:0046635 | positive regulation of alpha-beta T cell activation(GO:0046635) |
| 0.1 | 0.4 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.1 | 0.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 1.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.8 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.2 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 0.3 | GO:0098586 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.1 | 0.4 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.6 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.2 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.4 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.1 | 0.2 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.5 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.3 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) neurohypophysis development(GO:0021985) maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.0 | 0.2 | GO:0050951 | detection of temperature stimulus(GO:0016048) sensory perception of temperature stimulus(GO:0050951) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.0 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.3 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.3 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.2 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.1 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.0 | 0.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0032640 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.0 | 0.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.3 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.4 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.4 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.1 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.4 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0060155 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.0 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.8 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.3 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.5 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.2 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.4 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.4 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.1 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.0 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.6 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.3 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.0 | GO:0015824 | L-alanine transport(GO:0015808) proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.5 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 1.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.7 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.8 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.3 | 1.7 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.2 | 0.5 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.1 | 0.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.5 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.4 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.1 | 0.3 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.4 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.2 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 0.8 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 1.0 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.0 | 0.2 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.0 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.3 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 4.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.1 | GO:0031779 | melanocortin receptor binding(GO:0031779) |
| 0.0 | 0.1 | GO:0005549 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.0 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.1 | GO:0038187 | pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 0.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |