Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
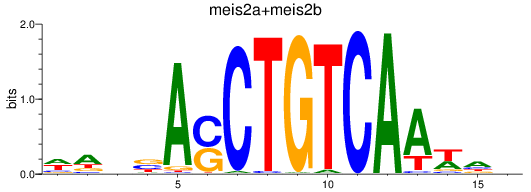
Results for meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2
Z-value: 0.57
Transcription factors associated with meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meis2b
|
ENSDARG00000077840 | Meis homeobox 2a |
|
meis2a
|
ENSDARG00000098240 | Meis homeobox 2a |
|
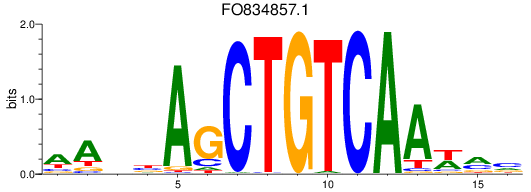
FO834857.1
|
ENSDARG00000112895 | homeobox protein TGIF2-like |
|
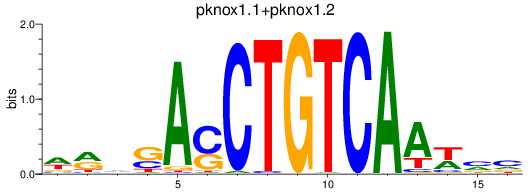
pknox1.1
|
ENSDARG00000018765 | pbx/knotted 1 homeobox 1.1 |
|
pknox1.2
|
ENSDARG00000036542 | pbx/knotted 1 homeobox 1.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meis2a | dr11_v1_chr17_+_52823015_52823138 | 0.86 | 2.4e-06 | Click! |
| pknox1.2 | dr11_v1_chr1_-_46981134_46981134 | 0.65 | 2.6e-03 | Click! |
| pknox1.1 | dr11_v1_chr9_+_19529951_19529951 | -0.48 | 3.9e-02 | Click! |
| meis2b | dr11_v1_chr20_-_10120442_10120442 | 0.22 | 3.7e-01 | Click! |
| FO834857.1 | dr11_v1_chr6_-_1814457_1814457 | 0.18 | 4.6e-01 | Click! |
Activity profile of meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2 motif
Sorted Z-values of meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22213297 | 4.47 |
ENSDART00000110656
ENSDART00000133149 |
crygm2d20
|
crystallin, gamma M2d20 |
| chr10_+_26800213 | 3.58 |
ENSDART00000078996
|
arr3a
|
arrestin 3a, retinal (X-arrestin) |
| chr7_+_10592152 | 3.19 |
ENSDART00000182624
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr13_-_7031033 | 2.83 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr15_+_19652807 | 2.33 |
ENSDART00000134321
ENSDART00000054426 |
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr1_-_22691182 | 2.03 |
ENSDART00000076766
|
fgfbp2b
|
fibroblast growth factor binding protein 2b |
| chr11_-_32723851 | 1.95 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr11_+_25459697 | 1.86 |
ENSDART00000161481
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr20_+_28434196 | 1.76 |
ENSDART00000034245
|
dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr8_+_34731982 | 1.62 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr14_+_46410766 | 1.46 |
ENSDART00000032342
|
anxa5a
|
annexin A5a |
| chr11_+_40649412 | 1.36 |
ENSDART00000043016
ENSDART00000134560 |
slc45a1
|
solute carrier family 45, member 1 |
| chr5_-_71722257 | 1.30 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr20_-_47731768 | 1.27 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr1_-_14234076 | 1.25 |
ENSDART00000040049
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr1_-_14233815 | 1.25 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr3_-_28428198 | 1.22 |
ENSDART00000151546
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr11_+_1796426 | 1.20 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr8_-_33114202 | 1.17 |
ENSDART00000098840
|
ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr8_+_25351863 | 1.13 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr4_+_7841627 | 1.09 |
ENSDART00000037997
|
ucmaa
|
upper zone of growth plate and cartilage matrix associated a |
| chr19_-_6134802 | 1.08 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr7_-_58269812 | 1.03 |
ENSDART00000050077
|
sdcbp
|
syndecan binding protein (syntenin) |
| chr3_-_36612877 | 0.98 |
ENSDART00000167164
|
si:dkeyp-72e1.7
|
si:dkeyp-72e1.7 |
| chr5_+_32246872 | 0.95 |
ENSDART00000139917
|
myha
|
myosin, heavy chain a |
| chr16_-_563235 | 0.94 |
ENSDART00000016303
|
irx2a
|
iroquois homeobox 2a |
| chr22_-_16377666 | 0.93 |
ENSDART00000161878
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr18_+_907266 | 0.93 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr12_-_22039350 | 0.91 |
ENSDART00000153187
|
thrab
|
thyroid hormone receptor alpha b |
| chr6_+_27151940 | 0.88 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr12_+_46960579 | 0.87 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr20_-_27733683 | 0.85 |
ENSDART00000103317
ENSDART00000138139 |
zgc:153157
|
zgc:153157 |
| chr23_+_5465806 | 0.85 |
ENSDART00000149434
ENSDART00000148506 |
tulp1a
|
tubby like protein 1a |
| chr1_-_44084071 | 0.83 |
ENSDART00000166912
|
vwa11
|
von Willebrand factor A domain containing 11 |
| chr13_+_45524475 | 0.82 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr3_+_13190012 | 0.82 |
ENSDART00000179747
ENSDART00000109876 ENSDART00000124824 ENSDART00000130261 |
sun1
|
Sad1 and UNC84 domain containing 1 |
| chr7_+_14291323 | 0.82 |
ENSDART00000053521
|
rhcga
|
Rh family, C glycoprotein a |
| chr5_+_27137473 | 0.81 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr6_+_38381957 | 0.81 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr13_+_46803979 | 0.81 |
ENSDART00000159260
|
CU695232.1
|
|
| chr20_-_43771871 | 0.80 |
ENSDART00000153304
|
matn3a
|
matrilin 3a |
| chr7_-_38340674 | 0.80 |
ENSDART00000075782
|
slc7a10a
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a |
| chr25_+_3326885 | 0.79 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr16_-_563732 | 0.73 |
ENSDART00000183394
|
irx2a
|
iroquois homeobox 2a |
| chr3_+_35298078 | 0.73 |
ENSDART00000110126
|
cacng3b
|
calcium channel, voltage-dependent, gamma subunit 3b |
| chr24_-_12981423 | 0.72 |
ENSDART00000133166
ENSDART00000131370 ENSDART00000002992 |
dcaf11
|
ddb1 and cul4 associated factor 11 |
| chr3_-_27880229 | 0.72 |
ENSDART00000151404
|
abat
|
4-aminobutyrate aminotransferase |
| chr21_+_45733871 | 0.72 |
ENSDART00000187285
ENSDART00000193018 |
zgc:77058
|
zgc:77058 |
| chr10_-_42131408 | 0.71 |
ENSDART00000076693
|
stambpa
|
STAM binding protein a |
| chr18_+_39487486 | 0.71 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr1_+_14253118 | 0.70 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr6_-_57539141 | 0.70 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr1_+_50997156 | 0.69 |
ENSDART00000010449
|
ugp2a
|
UDP-glucose pyrophosphorylase 2a |
| chr23_-_7826849 | 0.68 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr8_+_23703680 | 0.67 |
ENSDART00000141099
ENSDART00000135394 |
ppardb
|
peroxisome proliferator-activated receptor delta b |
| chr25_-_21894317 | 0.66 |
ENSDART00000089642
|
fbxo31
|
F-box protein 31 |
| chr2_-_39090258 | 0.66 |
ENSDART00000134270
ENSDART00000145268 |
NMNAT3
|
si:ch73-170l17.1 |
| chr24_+_42148140 | 0.66 |
ENSDART00000010658
|
serpinb1l3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 3 |
| chr21_-_41025340 | 0.65 |
ENSDART00000148231
|
plac8l1
|
PLAC8-like 1 |
| chr6_-_40697585 | 0.65 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr19_+_10396042 | 0.64 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr8_-_50525360 | 0.63 |
ENSDART00000175648
|
CABZ01060030.1
|
|
| chr24_-_37568359 | 0.63 |
ENSDART00000056286
|
h1f0
|
H1 histone family, member 0 |
| chr21_+_39336285 | 0.63 |
ENSDART00000139677
|
si:ch211-274p24.4
|
si:ch211-274p24.4 |
| chr16_+_50434668 | 0.62 |
ENSDART00000193500
|
IGLON5
|
zgc:110372 |
| chr13_+_36764715 | 0.62 |
ENSDART00000111832
ENSDART00000085230 |
atl1
|
atlastin GTPase 1 |
| chr19_-_10395683 | 0.61 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr11_+_38280454 | 0.60 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr9_-_48397702 | 0.60 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr11_+_44356504 | 0.59 |
ENSDART00000160678
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr7_-_71829649 | 0.58 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr13_+_7292061 | 0.58 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr23_-_18024543 | 0.58 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr5_-_13766651 | 0.57 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr4_-_15603511 | 0.56 |
ENSDART00000122520
ENSDART00000162356 |
chchd3a
|
coiled-coil-helix-coiled-coil-helix domain containing 3a |
| chr14_-_32255126 | 0.56 |
ENSDART00000017259
|
fgf13a
|
fibroblast growth factor 13a |
| chr5_-_69482891 | 0.55 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr6_+_48041759 | 0.54 |
ENSDART00000140086
|
si:dkey-92f12.2
|
si:dkey-92f12.2 |
| chr23_+_19558574 | 0.54 |
ENSDART00000137811
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr21_-_16399778 | 0.53 |
ENSDART00000136435
|
unc5da
|
unc-5 netrin receptor Da |
| chr25_+_3327071 | 0.52 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr9_-_12575776 | 0.52 |
ENSDART00000128931
ENSDART00000182695 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr3_+_22273123 | 0.52 |
ENSDART00000044157
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr21_+_38312549 | 0.52 |
ENSDART00000065159
|
zgc:158291
|
zgc:158291 |
| chr22_-_14128716 | 0.51 |
ENSDART00000140323
|
si:ch211-246m6.4
|
si:ch211-246m6.4 |
| chr5_-_25406807 | 0.51 |
ENSDART00000089748
|
rorb
|
RAR-related orphan receptor B |
| chr8_-_410728 | 0.51 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr7_-_38638276 | 0.49 |
ENSDART00000074463
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr8_+_23213320 | 0.49 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr9_-_12575569 | 0.48 |
ENSDART00000102419
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr22_+_7480465 | 0.48 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr15_+_21862295 | 0.48 |
ENSDART00000154449
ENSDART00000180774 |
si:dkey-103g5.3
|
si:dkey-103g5.3 |
| chr11_-_41132296 | 0.47 |
ENSDART00000162944
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr12_+_17504559 | 0.47 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr4_+_27898833 | 0.47 |
ENSDART00000146099
|
cerk
|
ceramide kinase |
| chr23_+_39963599 | 0.46 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr2_+_52232630 | 0.46 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr16_-_16152199 | 0.46 |
ENSDART00000012718
|
fabp11b
|
fatty acid binding protein 11b |
| chr20_+_14968031 | 0.46 |
ENSDART00000151805
ENSDART00000151448 ENSDART00000063874 ENSDART00000190910 |
vamp4
|
vesicle-associated membrane protein 4 |
| chr16_+_12240605 | 0.45 |
ENSDART00000060056
|
tpi1b
|
triosephosphate isomerase 1b |
| chr19_+_46824723 | 0.45 |
ENSDART00000158620
|
AL953858.1
|
|
| chr17_+_33999630 | 0.45 |
ENSDART00000167085
ENSDART00000155030 ENSDART00000168522 ENSDART00000191799 ENSDART00000189684 ENSDART00000153942 ENSDART00000187272 ENSDART00000127692 |
gphna
|
gephyrin a |
| chr15_-_34845414 | 0.45 |
ENSDART00000009892
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr21_-_32684570 | 0.44 |
ENSDART00000162873
|
adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr23_+_33963619 | 0.44 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr1_+_41666611 | 0.44 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr11_+_30513656 | 0.43 |
ENSDART00000008594
|
tmem178
|
transmembrane protein 178 |
| chr24_-_36095526 | 0.43 |
ENSDART00000158145
|
CABZ01075509.1
|
|
| chr21_+_1380099 | 0.43 |
ENSDART00000184516
|
TCF4
|
transcription factor 4 |
| chr25_-_3549321 | 0.43 |
ENSDART00000181214
ENSDART00000160600 |
hdhd5
|
haloacid dehalogenase like hydrolase domain containing 5 |
| chr8_-_17167819 | 0.42 |
ENSDART00000135042
ENSDART00000143920 |
mrps36
|
mitochondrial ribosomal protein S36 |
| chr23_-_15216654 | 0.42 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr24_+_3963684 | 0.42 |
ENSDART00000182959
ENSDART00000185926 ENSDART00000167043 ENSDART00000033394 |
pfkpa
|
phosphofructokinase, platelet a |
| chr12_-_43982343 | 0.42 |
ENSDART00000161539
|
CR385054.1
|
|
| chr15_-_46718759 | 0.42 |
ENSDART00000085926
ENSDART00000154577 |
zgc:162872
|
zgc:162872 |
| chr3_-_28258462 | 0.41 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr5_+_6796291 | 0.41 |
ENSDART00000166868
ENSDART00000165308 |
me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr5_-_18474486 | 0.41 |
ENSDART00000090580
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr23_+_21067684 | 0.41 |
ENSDART00000132066
|
kcnab2b
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 b |
| chr11_+_24001993 | 0.41 |
ENSDART00000168215
|
chia.2
|
chitinase, acidic.2 |
| chr18_+_507835 | 0.40 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr3_-_32169754 | 0.39 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr2_-_51794472 | 0.39 |
ENSDART00000186652
|
BX908782.3
|
|
| chr3_+_24986145 | 0.38 |
ENSDART00000055428
|
cbx7a
|
chromobox homolog 7a |
| chr23_-_36592436 | 0.38 |
ENSDART00000168246
|
spryd3
|
SPRY domain containing 3 |
| chr23_-_29667544 | 0.37 |
ENSDART00000059339
|
clstn1
|
calsyntenin 1 |
| chr20_-_25522911 | 0.37 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr22_+_19188809 | 0.37 |
ENSDART00000134791
ENSDART00000133682 |
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr2_+_39021282 | 0.37 |
ENSDART00000056577
|
RBP1 (1 of many)
|
si:ch211-119o8.7 |
| chr21_-_43474012 | 0.36 |
ENSDART00000065104
|
tmem185
|
transmembrane protein 185 |
| chr5_+_15992655 | 0.36 |
ENSDART00000182148
|
znrf3
|
zinc and ring finger 3 |
| chr5_+_19320554 | 0.36 |
ENSDART00000165119
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr14_+_29200772 | 0.35 |
ENSDART00000166608
|
TENM2
|
si:dkey-34l15.2 |
| chr11_+_6422374 | 0.35 |
ENSDART00000183148
|
FO681393.1
|
|
| chr6_+_23931236 | 0.35 |
ENSDART00000166079
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr2_+_5620742 | 0.35 |
ENSDART00000028390
|
fgf12a
|
fibroblast growth factor 12a |
| chr22_-_21897203 | 0.35 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr16_+_11151699 | 0.35 |
ENSDART00000140674
|
cicb
|
capicua transcriptional repressor b |
| chr17_+_37624704 | 0.34 |
ENSDART00000157001
|
tmem229b
|
transmembrane protein 229B |
| chr4_+_22613625 | 0.34 |
ENSDART00000131161
|
sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr10_-_20669635 | 0.34 |
ENSDART00000131361
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr10_-_7671219 | 0.34 |
ENSDART00000159330
|
pcyox1
|
prenylcysteine oxidase 1 |
| chr18_-_7810214 | 0.34 |
ENSDART00000139505
ENSDART00000139188 |
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr22_+_1006573 | 0.33 |
ENSDART00000123458
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr18_-_444168 | 0.33 |
ENSDART00000170389
|
wtip
|
WT1 interacting protein |
| chr2_+_9918935 | 0.33 |
ENSDART00000140434
|
acbd5b
|
acyl-CoA binding domain containing 5b |
| chr3_-_50146854 | 0.33 |
ENSDART00000157047
|
si:dkey-120c6.5
|
si:dkey-120c6.5 |
| chr1_+_45217425 | 0.33 |
ENSDART00000179983
ENSDART00000074683 |
EVI5L
|
si:ch211-239f4.1 |
| chr4_-_17629444 | 0.32 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr18_-_33344 | 0.32 |
ENSDART00000129125
|
pde8a
|
phosphodiesterase 8A |
| chr22_-_16377960 | 0.32 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr13_+_27314795 | 0.32 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr18_-_8313686 | 0.32 |
ENSDART00000182187
|
mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr9_-_22821901 | 0.32 |
ENSDART00000101711
|
neb
|
nebulin |
| chr22_-_23253481 | 0.32 |
ENSDART00000054807
|
lhx9
|
LIM homeobox 9 |
| chr2_+_9918678 | 0.31 |
ENSDART00000081253
|
acbd5b
|
acyl-CoA binding domain containing 5b |
| chr1_+_2301961 | 0.31 |
ENSDART00000108919
ENSDART00000143361 ENSDART00000142944 |
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr20_-_46467280 | 0.31 |
ENSDART00000060702
|
rmdn3
|
regulator of microtubule dynamics 3 |
| chr7_-_68373495 | 0.31 |
ENSDART00000167440
|
zfhx3
|
zinc finger homeobox 3 |
| chr4_-_5291256 | 0.31 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr5_-_323712 | 0.30 |
ENSDART00000188793
|
HOOK3
|
hook microtubule tethering protein 3 |
| chr16_-_20492799 | 0.30 |
ENSDART00000014769
|
chn2
|
chimerin 2 |
| chr12_+_32159272 | 0.30 |
ENSDART00000153167
|
hlfb
|
hepatic leukemia factor b |
| chr7_-_27685365 | 0.30 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr4_+_4267451 | 0.30 |
ENSDART00000192069
|
ano2
|
anoctamin 2 |
| chr23_-_4019699 | 0.30 |
ENSDART00000159780
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr21_+_45496442 | 0.29 |
ENSDART00000164880
|
si:dkey-223p19.1
|
si:dkey-223p19.1 |
| chr12_-_44171241 | 0.29 |
ENSDART00000169233
ENSDART00000165496 |
si:dkey-31i7.2
|
si:dkey-31i7.2 |
| chr20_+_3339248 | 0.29 |
ENSDART00000108955
|
mfsd4b
|
major facilitator superfamily domain containing 4B |
| chr20_-_39188476 | 0.29 |
ENSDART00000152858
|
rcan2
|
regulator of calcineurin 2 |
| chr8_-_18899427 | 0.29 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr10_+_466926 | 0.28 |
ENSDART00000145220
|
arvcfa
|
ARVCF, delta catenin family member a |
| chr2_+_394166 | 0.28 |
ENSDART00000155733
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr13_+_28417297 | 0.28 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr8_-_17067364 | 0.28 |
ENSDART00000132687
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr21_+_45627775 | 0.28 |
ENSDART00000185466
|
irf1b
|
interferon regulatory factor 1b |
| chr1_+_55137943 | 0.28 |
ENSDART00000138070
ENSDART00000150510 ENSDART00000133472 ENSDART00000136378 |
mb
|
myoglobin |
| chr21_-_20939488 | 0.28 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr8_+_25352268 | 0.28 |
ENSDART00000187829
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr2_-_12502495 | 0.27 |
ENSDART00000186781
|
dpp6b
|
dipeptidyl-peptidase 6b |
| chr17_+_35362851 | 0.27 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr6_-_49510553 | 0.27 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr13_-_11667661 | 0.27 |
ENSDART00000102411
|
dctn1b
|
dynactin 1b |
| chr13_+_32446169 | 0.27 |
ENSDART00000143325
|
nt5c1ba
|
5'-nucleotidase, cytosolic IB a |
| chr3_-_60316118 | 0.27 |
ENSDART00000171458
|
si:ch211-214b16.2
|
si:ch211-214b16.2 |
| chr7_+_10610791 | 0.27 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr12_+_29055143 | 0.27 |
ENSDART00000076322
|
gabrz
|
gamma-aminobutyric acid (GABA) A receptor, zeta |
| chr23_+_20408227 | 0.27 |
ENSDART00000134727
|
si:rp71-17i16.4
|
si:rp71-17i16.4 |
| chr17_+_52823015 | 0.27 |
ENSDART00000160507
ENSDART00000186979 |
meis2a
|
Meis homeobox 2a |
| chr16_-_3678976 | 0.27 |
ENSDART00000111823
|
GRIK3
|
glutamate ionotropic receptor kainate type subunit 3 |
| chr16_-_35329803 | 0.27 |
ENSDART00000161729
ENSDART00000157700 ENSDART00000184584 ENSDART00000174713 ENSDART00000162518 |
ptprub
|
protein tyrosine phosphatase, receptor type, U, b |
| chr1_+_57187794 | 0.27 |
ENSDART00000152485
|
si:dkey-27j5.9
|
si:dkey-27j5.9 |
| chr4_+_9612574 | 0.27 |
ENSDART00000150336
ENSDART00000041289 ENSDART00000150828 |
tmem243b
|
transmembrane protein 243, mitochondrial b |
| chr21_+_37445871 | 0.26 |
ENSDART00000076333
|
pgk1
|
phosphoglycerate kinase 1 |
| chr2_-_24369087 | 0.26 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr4_-_815871 | 0.26 |
ENSDART00000067455
|
dpysl5b
|
dihydropyrimidinase-like 5b |
| chr3_-_28750495 | 0.26 |
ENSDART00000054408
|
gsg1l
|
gsg1-like |
| chr20_-_54564018 | 0.26 |
ENSDART00000099832
|
zgc:153012
|
zgc:153012 |
| chr9_-_49493305 | 0.26 |
ENSDART00000148707
ENSDART00000148561 |
xirp2b
|
xin actin binding repeat containing 2b |
| chr18_-_49286381 | 0.26 |
ENSDART00000174248
ENSDART00000174038 |
si:zfos-464b6.2
|
si:zfos-464b6.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.3 | 3.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.3 | 0.8 | GO:0042942 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.2 | 0.7 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.2 | 0.6 | GO:0097376 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.2 | 1.3 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.2 | 0.7 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 0.8 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.4 | GO:0018315 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 1.5 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 0.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 0.6 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.7 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 0.7 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.7 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 0.7 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 1.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.5 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.3 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 0.6 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.1 | 0.8 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 1.0 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 0.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 1.7 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.1 | 0.8 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.5 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.3 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.1 | 0.4 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 0.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 1.4 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 1.1 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 0.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.1 | 0.6 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.1 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.4 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.2 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 1.8 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 1.9 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 0.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 1.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 1.6 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.7 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.2 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.3 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.4 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0055057 | neuroblast division(GO:0055057) |
| 0.0 | 0.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.0 | 0.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.3 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.1 | GO:0043084 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.4 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.1 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.0 | 1.4 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 3.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.8 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.2 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.3 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.1 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.4 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 0.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.6 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.1 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.0 | 0.1 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.0 | 0.2 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.8 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 1.1 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.1 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.6 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 1.0 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.4 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 2.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 1.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.5 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.5 | 1.6 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.3 | 1.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 0.7 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 1.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 0.7 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.2 | 1.4 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 0.5 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.2 | 0.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 1.4 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.7 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 1.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.5 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.4 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.3 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 1.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 6.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.3 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.1 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.2 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 1.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 2.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.5 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 1.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 1.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 2.3 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 1.2 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 3.9 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.0 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.2 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |