Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
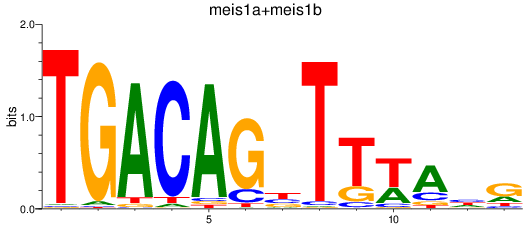
Results for meis1a+meis1b
Z-value: 1.36
Transcription factors associated with meis1a+meis1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meis1a
|
ENSDARG00000002937 | Meis homeobox 1 a |
|
meis1b
|
ENSDARG00000012078 | Meis homeobox 1 b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meis1a | dr11_v1_chr1_+_51496862_51496862 | -0.86 | 2.2e-06 | Click! |
| meis1b | dr11_v1_chr13_-_5569562_5569562 | -0.58 | 8.9e-03 | Click! |
Activity profile of meis1a+meis1b motif
Sorted Z-values of meis1a+meis1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_1796426 | 8.10 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr16_+_52512025 | 7.43 |
ENSDART00000056095
|
fabp10a
|
fatty acid binding protein 10a, liver basic |
| chr20_+_35473288 | 3.50 |
ENSDART00000047195
|
mep1a.1
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 1 |
| chr19_+_40856534 | 3.39 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr25_-_4482449 | 3.39 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr11_+_25459697 | 3.32 |
ENSDART00000161481
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr19_+_48111285 | 3.22 |
ENSDART00000169420
|
nme2b.2
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 2 |
| chr15_+_19652807 | 3.19 |
ENSDART00000134321
ENSDART00000054426 |
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr13_-_7031033 | 3.12 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr16_-_13612650 | 3.04 |
ENSDART00000080372
|
dbpb
|
D site albumin promoter binding protein b |
| chr19_+_40856807 | 3.03 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr24_+_3307857 | 2.95 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr1_+_14432434 | 2.82 |
ENSDART00000024328
|
slc34a2a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 2a |
| chr18_+_1703984 | 2.80 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr9_-_43071519 | 2.76 |
ENSDART00000109099
|
ttn.2
|
titin, tandem duplicate 2 |
| chr10_-_22831611 | 2.73 |
ENSDART00000160115
|
per1a
|
period circadian clock 1a |
| chr20_-_7176809 | 2.73 |
ENSDART00000012247
ENSDART00000125019 |
dhcr24
|
24-dehydrocholesterol reductase |
| chr20_-_5291012 | 2.68 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr16_-_16152199 | 2.66 |
ENSDART00000012718
|
fabp11b
|
fatty acid binding protein 11b |
| chr23_-_27875140 | 2.63 |
ENSDART00000143662
|
ankrd33aa
|
ankyrin repeat domain 33Aa |
| chr13_+_27314795 | 2.53 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr24_-_27400017 | 2.52 |
ENSDART00000145829
|
ccl34b.1
|
chemokine (C-C motif) ligand 34b, duplicate 1 |
| chr4_-_9549693 | 2.46 |
ENSDART00000160242
|
FQ377934.1
|
|
| chr10_-_22249444 | 2.39 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr19_-_6988837 | 2.37 |
ENSDART00000145741
ENSDART00000167640 |
znf384l
|
zinc finger protein 384 like |
| chr4_+_7841627 | 2.34 |
ENSDART00000037997
|
ucmaa
|
upper zone of growth plate and cartilage matrix associated a |
| chr17_+_22311413 | 2.33 |
ENSDART00000151929
|
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr6_-_8580857 | 2.31 |
ENSDART00000138858
ENSDART00000041142 |
myh11a
|
myosin, heavy chain 11a, smooth muscle |
| chr13_+_27073901 | 2.29 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr16_-_13613475 | 2.29 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr2_+_394166 | 2.27 |
ENSDART00000155733
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr14_+_22172047 | 2.25 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr20_-_1151265 | 2.24 |
ENSDART00000012376
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr20_-_17041025 | 2.22 |
ENSDART00000063764
|
si:dkey-5n18.1
|
si:dkey-5n18.1 |
| chr11_+_40649412 | 2.22 |
ENSDART00000043016
ENSDART00000134560 |
slc45a1
|
solute carrier family 45, member 1 |
| chr5_-_26093945 | 2.22 |
ENSDART00000010199
ENSDART00000145096 |
fam219ab
|
family with sequence similarity 219, member Ab |
| chr13_-_21701323 | 2.18 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr7_-_65492048 | 2.17 |
ENSDART00000162381
|
dkk3a
|
dickkopf WNT signaling pathway inhibitor 3a |
| chr24_-_11325849 | 2.16 |
ENSDART00000182485
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr18_+_28564640 | 2.16 |
ENSDART00000016983
|
spon1a
|
spondin 1a |
| chr21_-_16399778 | 2.13 |
ENSDART00000136435
|
unc5da
|
unc-5 netrin receptor Da |
| chr10_+_25726694 | 2.10 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr8_-_21268303 | 2.09 |
ENSDART00000067211
|
gpr37l1b
|
G protein-coupled receptor 37 like 1b |
| chr16_+_11558868 | 2.09 |
ENSDART00000112497
ENSDART00000180445 |
zgc:198329
|
zgc:198329 |
| chr5_+_26079178 | 2.08 |
ENSDART00000145920
|
si:dkey-201c13.2
|
si:dkey-201c13.2 |
| chr11_-_36957127 | 2.06 |
ENSDART00000168528
|
cacna1da
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, a |
| chr16_+_6944311 | 2.05 |
ENSDART00000144763
|
eaf1
|
ELL associated factor 1 |
| chr12_-_31461219 | 2.02 |
ENSDART00000148954
|
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr21_+_11468934 | 2.01 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr25_+_10458990 | 1.95 |
ENSDART00000130354
ENSDART00000044738 |
ric8a
|
RIC8 guanine nucleotide exchange factor A |
| chr7_+_73444325 | 1.93 |
ENSDART00000123016
|
si:ch211-142d6.2
|
si:ch211-142d6.2 |
| chr2_+_7192966 | 1.92 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr5_-_13086616 | 1.91 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr16_-_50175069 | 1.89 |
ENSDART00000192979
|
lim2.5
|
lens intrinsic membrane protein 2.5 |
| chr12_+_1286642 | 1.89 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr11_-_24428237 | 1.87 |
ENSDART00000189107
ENSDART00000103752 |
dvl1b
|
dishevelled segment polarity protein 1b |
| chr16_-_18960613 | 1.86 |
ENSDART00000183197
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr8_+_19621511 | 1.86 |
ENSDART00000017128
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr10_+_21576909 | 1.85 |
ENSDART00000168604
ENSDART00000166533 |
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr6_+_58280936 | 1.85 |
ENSDART00000155244
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr3_-_1190132 | 1.85 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr25_-_17403598 | 1.85 |
ENSDART00000104216
|
cyp2x9
|
cytochrome P450, family 2, subfamily X, polypeptide 9 |
| chr22_-_13851297 | 1.83 |
ENSDART00000080306
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr5_-_18446483 | 1.83 |
ENSDART00000180027
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr17_+_31185276 | 1.82 |
ENSDART00000062887
|
disp2
|
dispatched homolog 2 (Drosophila) |
| chr2_-_28671139 | 1.81 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr13_+_3819475 | 1.78 |
ENSDART00000139958
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr7_-_13884610 | 1.78 |
ENSDART00000006897
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr21_-_28523548 | 1.75 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr16_+_50089417 | 1.75 |
ENSDART00000153675
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr9_-_22822084 | 1.73 |
ENSDART00000142020
|
neb
|
nebulin |
| chr2_-_38125657 | 1.69 |
ENSDART00000143433
|
cbln12
|
cerebellin 12 |
| chr20_-_5267600 | 1.68 |
ENSDART00000099258
|
cyp46a1.4
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 4 |
| chr4_-_5291256 | 1.64 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr15_-_39963009 | 1.61 |
ENSDART00000157388
|
lypc
|
ly6 domain containing, pigment cell |
| chr7_+_34227075 | 1.61 |
ENSDART00000052477
ENSDART00000173732 |
lctla
|
lactase-like a |
| chr18_-_21271373 | 1.60 |
ENSDART00000060001
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr22_-_33679277 | 1.59 |
ENSDART00000169948
|
FO904977.1
|
|
| chr8_+_19621731 | 1.59 |
ENSDART00000144667
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr6_+_48041759 | 1.57 |
ENSDART00000140086
|
si:dkey-92f12.2
|
si:dkey-92f12.2 |
| chr21_-_44556148 | 1.55 |
ENSDART00000163955
|
brcc3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr6_+_3280939 | 1.55 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr8_-_49399470 | 1.55 |
ENSDART00000136395
|
si:ch211-194e1.7
|
si:ch211-194e1.7 |
| chr8_-_45838277 | 1.54 |
ENSDART00000046064
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr10_-_7974155 | 1.51 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr7_-_67214972 | 1.51 |
ENSDART00000156861
|
swap70a
|
switching B cell complex subunit SWAP70a |
| chr9_-_22821901 | 1.48 |
ENSDART00000101711
|
neb
|
nebulin |
| chr19_-_9503473 | 1.47 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr6_-_24203980 | 1.46 |
ENSDART00000170915
|
si:ch73-389b16.2
|
si:ch73-389b16.2 |
| chr11_-_9948487 | 1.46 |
ENSDART00000189677
ENSDART00000113171 |
nlgn1
|
neuroligin 1 |
| chr9_+_29643036 | 1.46 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr23_+_40410644 | 1.43 |
ENSDART00000056328
|
elovl4b
|
ELOVL fatty acid elongase 4b |
| chr8_+_52284075 | 1.39 |
ENSDART00000098439
|
ube2d4
|
ubiquitin-conjugating enzyme E2D 4 (putative) |
| chr5_-_21030934 | 1.39 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr16_-_18960333 | 1.39 |
ENSDART00000143185
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr3_-_1232384 | 1.38 |
ENSDART00000053892
|
si:ch1073-314i13.4
|
si:ch1073-314i13.4 |
| chr21_+_43178831 | 1.38 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr13_+_36770738 | 1.37 |
ENSDART00000146696
|
atl1
|
atlastin GTPase 1 |
| chr13_+_24584401 | 1.36 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr14_+_2243 | 1.34 |
ENSDART00000191193
|
CYTL1
|
cytokine like 1 |
| chr17_+_18117358 | 1.34 |
ENSDART00000144894
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr15_+_9294620 | 1.33 |
ENSDART00000133588
ENSDART00000140009 |
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr22_+_15336752 | 1.33 |
ENSDART00000139070
|
sult3st2
|
sulfotransferase family 3, cytosolic sulfotransferase 2 |
| chr8_-_17771755 | 1.32 |
ENSDART00000063592
|
prkcz
|
protein kinase C, zeta |
| chr8_+_22582146 | 1.31 |
ENSDART00000157655
ENSDART00000189892 |
CT583651.2
|
|
| chr5_+_31480342 | 1.31 |
ENSDART00000098197
|
si:dkey-220k22.1
|
si:dkey-220k22.1 |
| chr24_+_4373033 | 1.29 |
ENSDART00000133360
|
ccny
|
cyclin Y |
| chr23_+_14432323 | 1.27 |
ENSDART00000178553
|
CR388373.3
|
|
| chr17_-_50331351 | 1.26 |
ENSDART00000149294
|
otofb
|
otoferlin b |
| chr15_-_29598444 | 1.26 |
ENSDART00000154847
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr22_+_5682635 | 1.25 |
ENSDART00000140680
ENSDART00000131308 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr24_-_17047918 | 1.24 |
ENSDART00000020204
|
msrb2
|
methionine sulfoxide reductase B2 |
| chr13_-_37545487 | 1.23 |
ENSDART00000131269
|
syt16
|
synaptotagmin XVI |
| chr10_+_41765944 | 1.22 |
ENSDART00000171484
|
rnf34b
|
ring finger protein 34b |
| chr18_+_507835 | 1.21 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr19_-_40826024 | 1.20 |
ENSDART00000131471
|
calcr
|
calcitonin receptor |
| chr3_-_60401826 | 1.19 |
ENSDART00000144030
ENSDART00000160821 |
si:ch211-214b16.4
|
si:ch211-214b16.4 |
| chr9_+_27411502 | 1.18 |
ENSDART00000143994
|
si:dkey-193n17.9
|
si:dkey-193n17.9 |
| chr24_+_27268001 | 1.18 |
ENSDART00000122639
|
vipr2
|
vasoactive intestinal peptide receptor 2 |
| chr25_-_35542739 | 1.18 |
ENSDART00000097651
|
si:ch211-87j1.4
|
si:ch211-87j1.4 |
| chr7_+_6990510 | 1.17 |
ENSDART00000138948
|
ccs
|
copper chaperone for superoxide dismutase |
| chr2_-_50372467 | 1.17 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein like 2b |
| chr17_-_43533095 | 1.17 |
ENSDART00000125162
|
si:dkey-21a6.5
|
si:dkey-21a6.5 |
| chr5_-_43682930 | 1.17 |
ENSDART00000075017
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr12_+_2993523 | 1.16 |
ENSDART00000082297
|
lrrc45
|
leucine rich repeat containing 45 |
| chr23_+_23183449 | 1.16 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr4_-_5302866 | 1.16 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr7_+_27898091 | 1.15 |
ENSDART00000186451
ENSDART00000173510 |
tub
|
tubby bipartite transcription factor |
| chr3_+_29082267 | 1.14 |
ENSDART00000145615
|
cacna1i
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr25_+_5604512 | 1.14 |
ENSDART00000042781
|
plxnb2b
|
plexin b2b |
| chr24_-_20320480 | 1.12 |
ENSDART00000137366
|
map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr13_-_29505604 | 1.12 |
ENSDART00000110005
|
cdhr1a
|
cadherin-related family member 1a |
| chr10_-_26512742 | 1.12 |
ENSDART00000135951
|
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr15_-_29598679 | 1.11 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr1_-_5455498 | 1.11 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr10_+_21722892 | 1.10 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr8_+_14792830 | 1.10 |
ENSDART00000139972
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr1_+_57331813 | 1.10 |
ENSDART00000152440
ENSDART00000062841 |
epn3b
|
epsin 3b |
| chr23_+_20689255 | 1.09 |
ENSDART00000182420
|
usp21
|
ubiquitin specific peptidase 21 |
| chr23_+_5470967 | 1.09 |
ENSDART00000110522
ENSDART00000149050 |
tulp1a
|
tubby like protein 1a |
| chr13_-_20519001 | 1.09 |
ENSDART00000168955
|
gfra1a
|
gdnf family receptor alpha 1a |
| chr20_+_23173710 | 1.09 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr10_+_22782522 | 1.08 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr9_-_12575776 | 1.08 |
ENSDART00000128931
ENSDART00000182695 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr16_-_38001040 | 1.07 |
ENSDART00000133861
ENSDART00000138711 ENSDART00000143846 ENSDART00000146564 |
si:ch211-198c19.3
|
si:ch211-198c19.3 |
| chr2_-_33676494 | 1.07 |
ENSDART00000141192
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr3_+_12861158 | 1.06 |
ENSDART00000171237
|
kcnj2b
|
potassium inwardly-rectifying channel, subfamily J, member 2b |
| chr15_+_42933236 | 1.06 |
ENSDART00000167763
|
slc8a2b
|
solute carrier family 8 (sodium/calcium exchanger), member 2b |
| chr23_-_11128601 | 1.06 |
ENSDART00000131232
|
cntn3a.2
|
contactin 3a, tandem duplicate 2 |
| chr4_-_5302162 | 1.05 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr17_-_29771639 | 1.05 |
ENSDART00000086201
|
ush2a
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr18_-_7810214 | 1.04 |
ENSDART00000139505
ENSDART00000139188 |
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr5_-_67661102 | 1.04 |
ENSDART00000013605
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr6_+_58915889 | 1.02 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr23_-_36724575 | 1.02 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr8_-_31384607 | 1.01 |
ENSDART00000164134
ENSDART00000024872 |
creb3l3l
|
cAMP responsive element binding protein 3-like 3 like |
| chr4_-_17629444 | 1.00 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr9_-_12575569 | 0.99 |
ENSDART00000102419
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr3_-_30609659 | 0.99 |
ENSDART00000182516
ENSDART00000187047 ENSDART00000110597 |
syt3
|
synaptotagmin III |
| chr25_+_32525131 | 0.99 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr20_-_8094718 | 0.98 |
ENSDART00000122902
|
si:ch211-232i5.3
|
si:ch211-232i5.3 |
| chr15_-_20956384 | 0.98 |
ENSDART00000135770
|
tbcela
|
tubulin folding cofactor E-like a |
| chr2_+_33335911 | 0.98 |
ENSDART00000126135
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr2_+_30147504 | 0.97 |
ENSDART00000190947
|
kcnb2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr10_+_4806323 | 0.96 |
ENSDART00000161789
|
palm2
|
paralemmin 2 |
| chr13_+_15182149 | 0.96 |
ENSDART00000193644
ENSDART00000134421 ENSDART00000086281 |
mavs
|
mitochondrial antiviral signaling protein |
| chr13_-_29505946 | 0.95 |
ENSDART00000026679
|
cdhr1a
|
cadherin-related family member 1a |
| chr19_+_7567763 | 0.94 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr10_-_3295197 | 0.92 |
ENSDART00000109131
|
slc25a1b
|
slc25a1 solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1b |
| chr2_-_18830722 | 0.92 |
ENSDART00000165330
ENSDART00000165698 |
pbx1a
|
pre-B-cell leukemia homeobox 1a |
| chr6_+_39493864 | 0.90 |
ENSDART00000086263
|
mettl7a
|
methyltransferase like 7A |
| chr15_-_4415917 | 0.89 |
ENSDART00000062874
|
atp1b3b
|
ATPase Na+/K+ transporting subunit beta 3b |
| chr6_+_54154807 | 0.89 |
ENSDART00000061735
|
nudt3b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3b |
| chr2_+_6127593 | 0.89 |
ENSDART00000184007
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr21_-_19006631 | 0.88 |
ENSDART00000080269
ENSDART00000191682 |
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr23_-_29667544 | 0.87 |
ENSDART00000059339
|
clstn1
|
calsyntenin 1 |
| chr8_-_16464453 | 0.87 |
ENSDART00000098691
|
rnf11b
|
ring finger protein 11b |
| chr15_+_24588963 | 0.86 |
ENSDART00000155075
|
zgc:198241
|
zgc:198241 |
| chr16_-_8132742 | 0.86 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr2_-_51719439 | 0.85 |
ENSDART00000170385
|
tgm1l1
|
transglutaminase 1 like 1 |
| chr2_+_11670270 | 0.85 |
ENSDART00000100524
|
ftr01
|
finTRIM family, member 1 |
| chr23_-_764135 | 0.84 |
ENSDART00000010248
ENSDART00000183978 |
mitfb
|
microphthalmia-associated transcription factor b |
| chr22_+_35275468 | 0.84 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr24_+_39656834 | 0.84 |
ENSDART00000133942
ENSDART00000148089 ENSDART00000145075 |
stub1
|
STIP1 homology and U-Box containing protein 1 |
| chr13_-_48161568 | 0.83 |
ENSDART00000109469
ENSDART00000188052 ENSDART00000193446 ENSDART00000189509 ENSDART00000184810 |
golga4
|
golgin A4 |
| chr24_+_10039165 | 0.83 |
ENSDART00000144186
|
pou6f2
|
POU class 6 homeobox 2 |
| chr4_-_18954001 | 0.83 |
ENSDART00000144814
|
si:dkey-31f5.8
|
si:dkey-31f5.8 |
| chr21_+_37357578 | 0.83 |
ENSDART00000143621
|
nsd1b
|
nuclear receptor binding SET domain protein 1b |
| chr10_-_39011514 | 0.83 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr11_+_41540862 | 0.82 |
ENSDART00000173210
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr25_-_4733221 | 0.82 |
ENSDART00000172689
|
drd4a
|
dopamine receptor D4a |
| chr17_+_18117976 | 0.82 |
ENSDART00000123745
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr11_-_34480822 | 0.81 |
ENSDART00000129029
|
xxylt1
|
xyloside xylosyltransferase 1 |
| chr24_-_20808283 | 0.80 |
ENSDART00000143759
|
vipr1b
|
vasoactive intestinal peptide receptor 1b |
| chr5_-_52569014 | 0.80 |
ENSDART00000165616
|
mamdc2a
|
MAM domain containing 2a |
| chr25_+_336503 | 0.79 |
ENSDART00000160395
|
CU929262.1
|
|
| chr20_-_5328637 | 0.79 |
ENSDART00000075974
|
ism2b
|
isthmin 2b |
| chr17_+_12138715 | 0.79 |
ENSDART00000112888
|
SCCPDH (1 of many)
|
zgc:174379 |
| chr16_+_30002605 | 0.79 |
ENSDART00000160555
|
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr21_+_5888641 | 0.79 |
ENSDART00000091331
|
prodha
|
proline dehydrogenase (oxidase) 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of meis1a+meis1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:2000191 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.6 | 2.2 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.5 | 3.2 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.5 | 1.6 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.5 | 1.5 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.5 | 2.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.4 | 4.9 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 3.4 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.3 | 1.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 0.3 | GO:0001659 | temperature homeostasis(GO:0001659) |
| 0.3 | 1.9 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.3 | 1.3 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.2 | 1.0 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.2 | 1.9 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.2 | 1.2 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.2 | 0.7 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.2 | 0.9 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.2 | 3.4 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.2 | 1.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.2 | 0.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 3.7 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.2 | 3.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.2 | 0.8 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.2 | 2.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.2 | 1.0 | GO:0098586 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.2 | 3.4 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 2.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 2.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 0.9 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 1.2 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 1.7 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.2 | 2.8 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.2 | 3.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 1.2 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.2 | 0.8 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 2.3 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 1.0 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 1.0 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.9 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 1.6 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.1 | 0.8 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 2.1 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.1 | 1.5 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 2.7 | GO:0043153 | response to hydrogen peroxide(GO:0042542) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.8 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 2.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.8 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.3 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 1.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.3 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.1 | 0.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) embryonic digestive tract morphogenesis(GO:0048557) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 1.3 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) |
| 0.1 | 0.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.4 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 0.5 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 1.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 2.3 | GO:0030856 | regulation of epithelial cell differentiation(GO:0030856) |
| 0.1 | 0.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 3.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.8 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.1 | 0.8 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 0.4 | GO:1901490 | protein kinase D signaling(GO:0089700) regulation of lymphangiogenesis(GO:1901490) |
| 0.1 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 1.5 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 1.4 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.1 | 2.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 1.8 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.1 | 0.3 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 2.9 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.2 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.1 | 1.8 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 0.3 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.4 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 1.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 0.5 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.1 | 1.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.5 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.1 | 2.1 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 0.5 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.4 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 1.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.5 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 2.2 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 1.1 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 1.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 6.1 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.0 | 1.6 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 1.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.4 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 2.8 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.5 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 1.8 | GO:0035195 | gene silencing by miRNA(GO:0035195) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.5 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.0 | 0.8 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.6 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.7 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.4 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 1.0 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.4 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 3.0 | GO:0008202 | steroid metabolic process(GO:0008202) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.7 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.4 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.7 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 0.8 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.2 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.4 | GO:1902866 | regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 4.0 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.7 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 4.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K4 trimethylation(GO:0080182) histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 2.6 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 1.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 1.0 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.1 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.2 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.3 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) |
| 0.0 | 0.5 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.9 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.8 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.4 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.4 | 6.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 1.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 0.6 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 1.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 2.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 0.8 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.5 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.3 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 1.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.0 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 2.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.8 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.7 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 1.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 3.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.4 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 0.8 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 2.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 2.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 4.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 2.8 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 3.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.4 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 4.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 4.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) calcium channel complex(GO:0034704) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 2.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 1.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.6 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 2.9 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 2.5 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 1.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.9 | 5.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.5 | 2.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.5 | 1.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.5 | 1.4 | GO:0042806 | fucose binding(GO:0042806) |
| 0.4 | 2.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.4 | 2.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.4 | 1.2 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.3 | 3.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.3 | 2.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.3 | 2.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.3 | 1.2 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.3 | 6.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 3.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.3 | 2.2 | GO:0008515 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.3 | 3.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.3 | 2.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.3 | 2.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 0.7 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 1.7 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 2.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 0.8 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.2 | 2.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 0.9 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 1.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 0.9 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.2 | 0.9 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.2 | 4.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.2 | 2.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 2.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 1.6 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 1.8 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 3.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 2.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 1.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.3 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 1.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 2.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 2.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 2.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.6 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 1.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 3.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 2.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.8 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.6 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 1.3 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 5.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.8 | GO:0051879 | Hsp70 protein binding(GO:0030544) Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 1.5 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 2.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 2.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 2.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 1.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 1.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 10.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 3.2 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 1.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 3.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 2.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.5 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 10.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.9 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.6 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 0.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 2.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.3 | 1.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.3 | 2.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 5.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 1.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 4.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 3.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 0.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |