Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
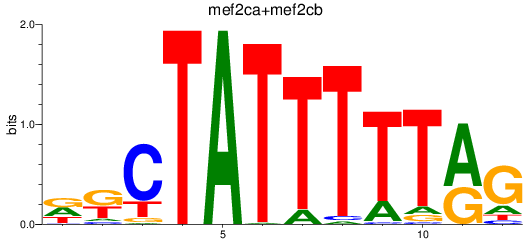
Results for mef2ca+mef2cb
Z-value: 1.09
Transcription factors associated with mef2ca+mef2cb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mef2cb
|
ENSDARG00000009418 | myocyte enhancer factor 2cb |
|
mef2ca
|
ENSDARG00000029764 | myocyte enhancer factor 2ca |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mef2cb | dr11_v1_chr5_-_48285756_48285756 | 0.72 | 5.0e-04 | Click! |
| mef2ca | dr11_v1_chr10_-_43611643_43611718 | 0.52 | 2.4e-02 | Click! |
Activity profile of mef2ca+mef2cb motif
Sorted Z-values of mef2ca+mef2cb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_45677781 | 4.60 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr25_+_31222069 | 4.39 |
ENSDART00000159373
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr3_-_61205711 | 4.27 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr24_-_4765740 | 3.86 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr14_+_32838110 | 3.78 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr6_-_12588044 | 3.41 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr12_-_4028079 | 3.22 |
ENSDART00000128676
|
si:ch211-180a12.2
|
si:ch211-180a12.2 |
| chr21_+_11684830 | 3.14 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr5_+_51597677 | 3.09 |
ENSDART00000048210
ENSDART00000184797 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr2_+_16536453 | 2.94 |
ENSDART00000100287
|
grk7a
|
G protein-coupled receptor kinase 7a |
| chr9_-_52814204 | 2.91 |
ENSDART00000140771
ENSDART00000007401 |
MAP3K13
|
si:ch211-45c16.2 |
| chr8_+_52637507 | 2.87 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr25_+_14092871 | 2.83 |
ENSDART00000067239
|
guca1g
|
guanylate cyclase activator 1g |
| chr14_+_32837914 | 2.69 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr1_-_43915423 | 2.68 |
ENSDART00000181915
ENSDART00000113673 |
scpp5
|
secretory calcium-binding phosphoprotein 5 |
| chr8_+_34731982 | 2.58 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr11_-_43104475 | 2.55 |
ENSDART00000125368
|
acyp2
|
acylphosphatase 2, muscle type |
| chr21_+_11685009 | 2.51 |
ENSDART00000014668
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr17_-_12336987 | 2.46 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr25_+_29160102 | 2.36 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr7_+_15266093 | 2.34 |
ENSDART00000124676
|
sv2ba
|
synaptic vesicle glycoprotein 2Ba |
| chr1_+_8601935 | 2.27 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr7_-_71758307 | 2.15 |
ENSDART00000161067
ENSDART00000165253 |
myom1b
|
myomesin 1b |
| chr16_-_43026273 | 2.10 |
ENSDART00000156820
ENSDART00000189080 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr10_-_22845485 | 2.08 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr3_+_26135502 | 2.05 |
ENSDART00000146979
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr16_-_19890303 | 2.04 |
ENSDART00000147161
ENSDART00000079159 |
hdac9b
|
histone deacetylase 9b |
| chr2_-_43168292 | 2.01 |
ENSDART00000132588
|
crema
|
cAMP responsive element modulator a |
| chr13_-_2189761 | 1.98 |
ENSDART00000166255
|
mlip
|
muscular LMNA-interacting protein |
| chr9_-_42873700 | 1.92 |
ENSDART00000125953
|
ttn.1
|
titin, tandem duplicate 1 |
| chr10_-_41450367 | 1.92 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr25_+_16945348 | 1.89 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr10_-_24371312 | 1.85 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr18_+_7264961 | 1.84 |
ENSDART00000188461
|
CABZ01015105.1
|
|
| chr21_+_12010505 | 1.84 |
ENSDART00000123522
|
aqp7
|
aquaporin 7 |
| chr3_-_49504023 | 1.82 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr20_+_13175379 | 1.81 |
ENSDART00000025644
|
ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha isoform |
| chr10_-_24362775 | 1.75 |
ENSDART00000182104
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr23_+_17220986 | 1.73 |
ENSDART00000054761
|
nol4lb
|
nucleolar protein 4-like b |
| chr6_-_38816500 | 1.65 |
ENSDART00000190866
ENSDART00000104124 |
cnga3a
|
cyclic nucleotide gated channel alpha 3a |
| chr15_-_9031996 | 1.64 |
ENSDART00000124998
|
rtn2a
|
reticulon 2a |
| chr22_+_17205608 | 1.64 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr7_+_41295974 | 1.59 |
ENSDART00000173568
ENSDART00000173544 |
si:dkey-86l18.10
|
si:dkey-86l18.10 |
| chr11_+_6819050 | 1.59 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr12_-_17707449 | 1.59 |
ENSDART00000142427
ENSDART00000034914 |
pvalb3
|
parvalbumin 3 |
| chr23_+_6077503 | 1.57 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr6_-_35472923 | 1.56 |
ENSDART00000185907
|
rgs8
|
regulator of G protein signaling 8 |
| chr20_-_39271844 | 1.56 |
ENSDART00000192708
|
clu
|
clusterin |
| chr1_-_8101495 | 1.52 |
ENSDART00000161938
|
si:dkeyp-9d4.3
|
si:dkeyp-9d4.3 |
| chr10_+_29698467 | 1.52 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr4_+_5180650 | 1.51 |
ENSDART00000067390
|
fgf6b
|
fibroblast growth factor 6b |
| chr18_+_40355408 | 1.50 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr17_-_50233493 | 1.49 |
ENSDART00000172266
|
fosaa
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
| chr16_-_17541890 | 1.49 |
ENSDART00000131328
|
clcn1b
|
chloride channel, voltage-sensitive 1b |
| chr1_+_19708508 | 1.49 |
ENSDART00000054581
ENSDART00000131206 |
march1
|
membrane-associated ring finger (C3HC4) 1 |
| chr20_+_23173710 | 1.48 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr7_-_41964877 | 1.45 |
ENSDART00000092351
ENSDART00000193395 ENSDART00000187947 |
neto2b
|
neuropilin (NRP) and tolloid (TLL)-like 2b |
| chr25_+_35212919 | 1.45 |
ENSDART00000180127
|
ano5a
|
anoctamin 5a |
| chr12_-_13886952 | 1.44 |
ENSDART00000110503
|
adam11
|
ADAM metallopeptidase domain 11 |
| chr3_+_23092762 | 1.43 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr6_+_11250033 | 1.42 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr3_+_59851537 | 1.42 |
ENSDART00000180997
|
CU693479.1
|
|
| chr5_+_62356304 | 1.41 |
ENSDART00000148381
|
aspa
|
aspartoacylase |
| chr11_+_25430851 | 1.41 |
ENSDART00000164999
ENSDART00000126403 |
si:dkey-13a21.4
|
si:dkey-13a21.4 |
| chr2_+_6181383 | 1.40 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr14_+_23811808 | 1.40 |
ENSDART00000014411
|
kctd16a
|
potassium channel tetramerization domain containing 16a |
| chr3_-_30123113 | 1.39 |
ENSDART00000153562
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr23_-_7826849 | 1.39 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr25_+_14165447 | 1.39 |
ENSDART00000145387
|
shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr5_-_10946232 | 1.37 |
ENSDART00000163139
ENSDART00000031265 |
rtn4r
|
reticulon 4 receptor |
| chr16_+_25245857 | 1.37 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr3_+_1735214 | 1.37 |
ENSDART00000185454
|
BX321875.3
|
|
| chr7_-_13381129 | 1.34 |
ENSDART00000164326
|
si:ch73-119p20.1
|
si:ch73-119p20.1 |
| chr4_+_11375894 | 1.34 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr17_-_43558494 | 1.33 |
ENSDART00000103830
|
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr3_+_28953274 | 1.32 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr7_-_42206720 | 1.30 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr17_-_50234004 | 1.30 |
ENSDART00000058706
|
fosaa
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
| chr13_-_11536951 | 1.28 |
ENSDART00000018155
|
adss
|
adenylosuccinate synthase |
| chr13_-_27767330 | 1.28 |
ENSDART00000131631
ENSDART00000112553 ENSDART00000189911 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr6_-_49547680 | 1.27 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr9_-_43073960 | 1.27 |
ENSDART00000059460
|
ttn.2
|
titin, tandem duplicate 2 |
| chr25_-_12809361 | 1.27 |
ENSDART00000162750
|
ca5a
|
carbonic anhydrase Va |
| chr23_-_18030399 | 1.27 |
ENSDART00000136967
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr15_-_34845414 | 1.26 |
ENSDART00000009892
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr8_-_32497815 | 1.25 |
ENSDART00000122359
|
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr23_-_35347714 | 1.25 |
ENSDART00000161770
ENSDART00000165615 |
cpne9
|
copine family member IX |
| chr7_-_71758613 | 1.24 |
ENSDART00000166724
|
myom1b
|
myomesin 1b |
| chr14_-_27297123 | 1.23 |
ENSDART00000173423
|
pcdh11
|
protocadherin 11 |
| chr10_-_2522588 | 1.23 |
ENSDART00000081926
|
CU856539.1
|
|
| chr9_+_29643036 | 1.23 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr3_-_49138004 | 1.22 |
ENSDART00000167173
|
gipc1
|
GIPC PDZ domain containing family, member 1 |
| chr18_-_25177230 | 1.21 |
ENSDART00000013363
|
slco3a1
|
solute carrier organic anion transporter family, member 3A1 |
| chr15_-_29162193 | 1.21 |
ENSDART00000138449
ENSDART00000099885 |
xaf1
|
XIAP associated factor 1 |
| chr23_-_27633730 | 1.21 |
ENSDART00000103639
|
arf3a
|
ADP-ribosylation factor 3a |
| chr19_+_24394560 | 1.20 |
ENSDART00000142506
|
si:dkey-81h8.1
|
si:dkey-81h8.1 |
| chr23_-_7125494 | 1.18 |
ENSDART00000111929
|
slco4a1
|
solute carrier organic anion transporter family, member 4A1 |
| chr3_-_14103551 | 1.15 |
ENSDART00000127604
|
nfil3-6
|
nuclear factor, interleukin 3 regulated, member 6 |
| chr21_-_25685739 | 1.15 |
ENSDART00000129619
ENSDART00000101205 |
phkg1b
|
phosphorylase kinase, gamma 1b (muscle) |
| chr23_+_17981127 | 1.15 |
ENSDART00000012571
ENSDART00000145200 |
chia.6
|
chitinase, acidic.6 |
| chr7_-_31321027 | 1.14 |
ENSDART00000186878
|
CR356242.1
|
|
| chr6_+_8079974 | 1.14 |
ENSDART00000152071
|
si:ch211-207j7.2
|
si:ch211-207j7.2 |
| chr1_-_49250490 | 1.14 |
ENSDART00000150386
|
si:ch73-6k14.2
|
si:ch73-6k14.2 |
| chr16_-_17188294 | 1.13 |
ENSDART00000165883
|
opn9
|
opsin 9 |
| chr13_+_22249636 | 1.13 |
ENSDART00000108472
ENSDART00000173123 |
synpo2la
|
synaptopodin 2-like a |
| chr9_+_22632126 | 1.13 |
ENSDART00000139434
|
etv5a
|
ets variant 5a |
| chr5_-_29122834 | 1.13 |
ENSDART00000087197
|
whrnb
|
whirlin b |
| chr15_+_40008370 | 1.12 |
ENSDART00000063783
|
itm2ca
|
integral membrane protein 2Ca |
| chr23_-_38497705 | 1.11 |
ENSDART00000109493
|
tshz2
|
teashirt zinc finger homeobox 2 |
| chr6_+_11250316 | 1.11 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr10_+_22724059 | 1.10 |
ENSDART00000136123
|
kdm6bb
|
lysine (K)-specific demethylase 6B, b |
| chr3_-_5016893 | 1.09 |
ENSDART00000165968
|
CSDC2 (1 of many)
|
cold shock domain containing C2 |
| chr8_+_30456161 | 1.09 |
ENSDART00000085894
|
pgm5
|
phosphoglucomutase 5 |
| chr8_-_49935358 | 1.08 |
ENSDART00000159782
ENSDART00000156841 |
agtpbp1
CU694202.1
|
ATP/GTP binding protein 1 |
| chr15_+_20543770 | 1.07 |
ENSDART00000092357
|
sgsm2
|
small G protein signaling modulator 2 |
| chr20_-_7000225 | 1.06 |
ENSDART00000100098
|
adcy1a
|
adenylate cyclase 1a |
| chr18_+_23193820 | 1.05 |
ENSDART00000148106
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr16_-_43356018 | 1.05 |
ENSDART00000181683
|
FO704821.1
|
|
| chr17_-_39886628 | 1.04 |
ENSDART00000002217
|
zmp:0000000545
|
zmp:0000000545 |
| chr20_+_18163821 | 1.03 |
ENSDART00000186507
|
aqp4
|
aquaporin 4 |
| chr21_+_43559123 | 1.02 |
ENSDART00000151212
|
gpr185a
|
G protein-coupled receptor 185 a |
| chr10_+_41765616 | 1.01 |
ENSDART00000170682
|
rnf34b
|
ring finger protein 34b |
| chr16_+_21738194 | 1.00 |
ENSDART00000163688
|
FP085428.1
|
Danio rerio si:ch211-154o6.4 (si:ch211-154o6.4), mRNA. |
| chr4_-_4119396 | 0.98 |
ENSDART00000067409
ENSDART00000138221 |
lmod2b
|
leiomodin 2 (cardiac) b |
| chr24_+_34085940 | 0.97 |
ENSDART00000171189
|
asb10
|
ankyrin repeat and SOCS box containing 10 |
| chr12_+_1000323 | 0.97 |
ENSDART00000054363
|
si:ch1073-272o11.3
|
si:ch1073-272o11.3 |
| chr17_-_722218 | 0.97 |
ENSDART00000160385
|
SLC25A29
|
solute carrier family 25 member 29 |
| chr21_+_9997418 | 0.97 |
ENSDART00000181454
ENSDART00000171579 |
herc7
|
hect domain and RLD 7 |
| chr6_+_11438972 | 0.96 |
ENSDART00000029314
|
col5a2b
|
collagen, type V, alpha 2b |
| chr2_+_6885852 | 0.96 |
ENSDART00000016607
|
rgs5b
|
regulator of G protein signaling 5b |
| chr7_+_13830052 | 0.95 |
ENSDART00000191360
|
abhd2a
|
abhydrolase domain containing 2a |
| chr9_-_23891102 | 0.95 |
ENSDART00000186799
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr2_-_37353098 | 0.95 |
ENSDART00000056522
|
skila
|
SKI-like proto-oncogene a |
| chr11_+_25634041 | 0.93 |
ENSDART00000033657
|
grm6b
|
glutamate receptor, metabotropic 6b |
| chr15_+_32867420 | 0.93 |
ENSDART00000159442
|
dclk1b
|
doublecortin-like kinase 1b |
| chr19_+_2590182 | 0.93 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr23_+_17980875 | 0.92 |
ENSDART00000163452
|
chia.6
|
chitinase, acidic.6 |
| chr4_-_72080351 | 0.92 |
ENSDART00000174925
|
LO017820.1
|
|
| chr11_+_11201096 | 0.92 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr3_-_42981739 | 0.91 |
ENSDART00000167844
|
mafk
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr11_-_19650781 | 0.91 |
ENSDART00000165595
|
SYNPR
|
si:dkey-30j16.3 |
| chr9_+_45227028 | 0.91 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr22_-_8509215 | 0.90 |
ENSDART00000140146
|
si:ch73-27e22.3
|
si:ch73-27e22.3 |
| chr21_+_40944530 | 0.90 |
ENSDART00000022976
|
kctd16b
|
potassium channel tetramerization domain containing 16b |
| chr5_+_22264051 | 0.90 |
ENSDART00000143314
|
arhgap20b
|
Rho GTPase activating protein 20b |
| chr16_-_22683038 | 0.89 |
ENSDART00000138130
|
s100t
|
S100 calcium binding protein T |
| chr12_-_26064480 | 0.88 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr3_-_30609659 | 0.88 |
ENSDART00000182516
ENSDART00000187047 ENSDART00000110597 |
syt3
|
synaptotagmin III |
| chr14_+_11909966 | 0.88 |
ENSDART00000171829
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr16_-_4026265 | 0.88 |
ENSDART00000149339
|
si:ch211-175f12.2
|
si:ch211-175f12.2 |
| chr19_-_9662958 | 0.88 |
ENSDART00000041094
|
clcn1a
|
chloride channel, voltage-sensitive 1a |
| chr21_+_40589770 | 0.88 |
ENSDART00000164650
ENSDART00000161584 ENSDART00000161108 |
pdk3b
|
pyruvate dehydrogenase kinase, isozyme 3b |
| chr12_+_32323098 | 0.87 |
ENSDART00000188722
|
ANKFN1
|
si:ch211-277e21.2 |
| chr1_+_50968908 | 0.87 |
ENSDART00000150353
ENSDART00000012842 |
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr20_+_16881883 | 0.86 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr24_-_21490628 | 0.86 |
ENSDART00000181546
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr3_+_44947355 | 0.86 |
ENSDART00000083040
|
hs3st2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
| chr11_+_11200550 | 0.85 |
ENSDART00000181339
ENSDART00000187116 |
myom2a
|
myomesin 2a |
| chr12_+_47794089 | 0.84 |
ENSDART00000160726
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr2_-_8611675 | 0.83 |
ENSDART00000138223
|
si:ch211-71m22.1
|
si:ch211-71m22.1 |
| chr1_-_9195629 | 0.83 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr20_+_34455645 | 0.83 |
ENSDART00000135789
|
mettl11b
|
methyltransferase like 11B |
| chr7_+_69356661 | 0.83 |
ENSDART00000177049
|
CABZ01083862.1
|
|
| chr10_+_40836378 | 0.83 |
ENSDART00000085792
|
trim69
|
tripartite motif containing 69 |
| chr11_-_1948784 | 0.82 |
ENSDART00000082475
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr10_+_37145007 | 0.82 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr1_+_31113951 | 0.82 |
ENSDART00000129362
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr8_+_22930627 | 0.81 |
ENSDART00000187860
|
sypa
|
synaptophysin a |
| chr7_+_42206847 | 0.79 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr13_+_31172833 | 0.79 |
ENSDART00000176378
|
CR931802.3
|
|
| chr5_-_29122615 | 0.78 |
ENSDART00000144802
|
whrnb
|
whirlin b |
| chr14_+_30291611 | 0.78 |
ENSDART00000173004
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr4_+_6572364 | 0.78 |
ENSDART00000122574
|
ppp1r3aa
|
protein phosphatase 1, regulatory subunit 3Aa |
| chr7_-_7420301 | 0.77 |
ENSDART00000102620
|
six7
|
SIX homeobox 7 |
| chr19_-_12648122 | 0.77 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr20_+_41549200 | 0.77 |
ENSDART00000135715
|
fam184a
|
family with sequence similarity 184, member A |
| chr18_+_23193567 | 0.77 |
ENSDART00000190072
ENSDART00000171594 ENSDART00000181762 |
mef2aa
|
myocyte enhancer factor 2aa |
| chr10_-_2527342 | 0.76 |
ENSDART00000184168
|
CU856539.1
|
|
| chr1_-_1885516 | 0.76 |
ENSDART00000122187
ENSDART00000131675 |
si:ch211-132g1.3
|
si:ch211-132g1.3 |
| chr18_-_8579907 | 0.76 |
ENSDART00000147284
|
FRMD4A
|
si:ch211-220f12.1 |
| chr8_+_47571211 | 0.76 |
ENSDART00000131460
|
plch2a
|
phospholipase C, eta 2a |
| chr1_-_38813679 | 0.75 |
ENSDART00000148917
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr18_-_12416019 | 0.75 |
ENSDART00000144799
|
si:ch211-1e14.1
|
si:ch211-1e14.1 |
| chr6_-_14139503 | 0.75 |
ENSDART00000089577
|
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr23_-_1557195 | 0.74 |
ENSDART00000136436
|
epm2a
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr1_+_7546259 | 0.74 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr25_-_31118923 | 0.74 |
ENSDART00000009126
ENSDART00000188286 |
kras
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| chr5_-_25236340 | 0.73 |
ENSDART00000162774
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr3_+_59051503 | 0.72 |
ENSDART00000160767
|
rasd4
|
rasd family member 4 |
| chr13_+_12175724 | 0.72 |
ENSDART00000166053
|
gabrg1
|
gamma-aminobutyric acid type A receptor gamma1 subunit |
| chr6_-_30859656 | 0.72 |
ENSDART00000156235
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr20_-_40360571 | 0.72 |
ENSDART00000144768
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr16_+_5259886 | 0.71 |
ENSDART00000186668
|
plecb
|
plectin b |
| chr17_-_50374005 | 0.71 |
ENSDART00000149773
|
otofb
|
otoferlin b |
| chr8_-_32497581 | 0.69 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr18_+_23218980 | 0.69 |
ENSDART00000185014
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr9_+_38088331 | 0.68 |
ENSDART00000123749
|
cacnb4a
|
calcium channel, voltage-dependent, beta 4a subunit |
| chr11_-_18254 | 0.68 |
ENSDART00000167814
|
prr13
|
proline rich 13 |
| chr25_-_8160030 | 0.67 |
ENSDART00000067159
|
tph1a
|
tryptophan hydroxylase 1 (tryptophan 5-monooxygenase) a |
| chr21_+_15870752 | 0.67 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
Network of associatons between targets according to the STRING database.
First level regulatory network of mef2ca+mef2cb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.5 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.5 | 1.6 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.5 | 2.6 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.5 | 2.9 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.4 | 2.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.4 | 1.6 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.4 | 3.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.4 | 5.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.4 | 1.5 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.3 | 1.4 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.3 | 1.3 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.3 | 0.9 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.3 | 1.1 | GO:0042985 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.3 | 1.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.3 | 2.5 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 2.9 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 1.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.2 | 1.8 | GO:0071331 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.2 | 3.2 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.2 | 1.0 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.2 | 2.1 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.2 | 0.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 1.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 2.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 0.9 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.2 | 0.8 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.2 | 3.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 2.3 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 1.0 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 2.1 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.4 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 2.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.4 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 1.3 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 3.4 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 1.0 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.4 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.7 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 1.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 1.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 4.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.3 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 1.3 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.1 | 1.9 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.4 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 1.8 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 2.6 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 0.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 4.5 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 0.3 | GO:0035521 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.3 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 2.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 1.4 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 1.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.7 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 1.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.5 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 0.5 | GO:0002698 | negative regulation of immune effector process(GO:0002698) |
| 0.0 | 0.8 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.5 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 2.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.2 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.4 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 1.8 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.4 | GO:0009583 | detection of light stimulus(GO:0009583) |
| 0.0 | 2.4 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 3.8 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 3.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 3.3 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 3.2 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.3 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:1901072 | glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 1.1 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.3 | GO:0033151 | V(D)J recombination(GO:0033151) |
| 0.0 | 0.2 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.1 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.0 | 0.4 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.6 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 1.3 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 1.0 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.9 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.8 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.4 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.5 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 1.3 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 1.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 2.3 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.6 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.4 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.4 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.6 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 1.4 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.6 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.6 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.0 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) |
| 0.0 | 1.1 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.2 | GO:0036294 | cellular response to decreased oxygen levels(GO:0036294) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.6 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 1.7 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.2 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0031673 | H zone(GO:0031673) |
| 0.4 | 2.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.3 | 0.8 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 1.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 1.9 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.2 | 4.4 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 1.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 3.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 1.9 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 1.3 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.2 | 1.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 2.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.4 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 1.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 4.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.2 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 2.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 1.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.3 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.3 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.1 | 1.3 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.1 | 1.0 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 1.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 3.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 2.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.7 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.9 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 2.7 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.9 | 2.6 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.6 | 2.9 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.5 | 3.6 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.5 | 2.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.5 | 3.4 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.4 | 2.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.4 | 1.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 2.8 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.3 | 2.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.2 | 1.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 1.8 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.2 | 2.9 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 0.6 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 2.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 4.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 3.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 0.9 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 1.0 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.2 | 1.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.4 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 0.7 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 1.0 | GO:0005372 | water transmembrane transporter activity(GO:0005372) water channel activity(GO:0015250) |
| 0.1 | 0.4 | GO:0019767 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.1 | 2.0 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.6 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.7 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 2.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 5.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.9 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 1.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 1.7 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 1.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.5 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 2.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 3.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.3 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 1.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.8 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.4 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.1 | 0.4 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 0.3 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 1.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 2.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.3 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.1 | 1.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.5 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.4 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.1 | GO:0042805 | actinin binding(GO:0042805) muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 2.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 2.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.5 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 1.1 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 1.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 6.1 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 1.5 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 4.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 5.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 0.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 1.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.7 | 2.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.6 | 5.6 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.3 | 2.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 4.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 1.8 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 0.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 1.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |