Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
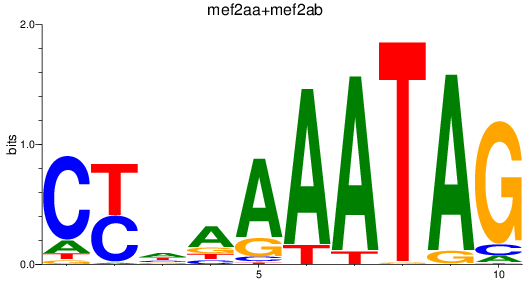
Results for mef2aa+mef2ab
Z-value: 2.16
Transcription factors associated with mef2aa+mef2ab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mef2aa
|
ENSDARG00000031756 | myocyte enhancer factor 2aa |
|
mef2ab
|
ENSDARG00000057527 | myocyte enhancer factor 2ab |
|
mef2ab
|
ENSDARG00000110771 | myocyte enhancer factor 2ab |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mef2aa | dr11_v1_chr18_+_23249519_23249519 | 0.93 | 1.2e-08 | Click! |
| mef2ab | dr11_v1_chr7_+_15659280_15659280 | 0.54 | 1.6e-02 | Click! |
Activity profile of mef2aa+mef2ab motif
Sorted Z-values of mef2aa+mef2ab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_61205711 | 15.47 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr9_-_22205682 | 12.57 |
ENSDART00000101869
|
crygm2d12
|
crystallin, gamma M2d12 |
| chr8_-_11112058 | 12.09 |
ENSDART00000042755
|
ampd1
|
adenosine monophosphate deaminase 1 (isoform M) |
| chr9_-_22188117 | 9.68 |
ENSDART00000132890
|
crygm2d17
|
crystallin, gamma M2d17 |
| chr5_+_45677781 | 9.28 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr5_+_51597677 | 9.16 |
ENSDART00000048210
ENSDART00000184797 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr12_-_4781801 | 8.61 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr8_+_34731982 | 7.68 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr7_-_27686021 | 6.76 |
ENSDART00000079112
ENSDART00000100989 |
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr3_+_32526263 | 6.38 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr25_+_19105804 | 6.04 |
ENSDART00000104414
|
rlbp1b
|
retinaldehyde binding protein 1b |
| chr12_-_17712393 | 6.02 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr16_-_36798783 | 5.91 |
ENSDART00000145697
|
calb1
|
calbindin 1 |
| chr3_+_59051503 | 5.87 |
ENSDART00000160767
|
rasd4
|
rasd family member 4 |
| chr4_-_16853464 | 5.87 |
ENSDART00000125743
ENSDART00000164570 |
slc25a3a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3a |
| chr15_+_19652807 | 5.66 |
ENSDART00000134321
ENSDART00000054426 |
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr12_-_4028079 | 5.52 |
ENSDART00000128676
|
si:ch211-180a12.2
|
si:ch211-180a12.2 |
| chr8_+_22930627 | 5.27 |
ENSDART00000187860
|
sypa
|
synaptophysin a |
| chr5_+_36932718 | 5.21 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr4_+_9279784 | 5.11 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr10_-_22845485 | 4.96 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr3_+_40170216 | 4.86 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr6_-_13891727 | 4.77 |
ENSDART00000065356
|
desmb
|
desmin b |
| chr1_+_8601935 | 4.76 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr13_-_2215213 | 4.65 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr2_-_5356686 | 4.40 |
ENSDART00000124290
|
MFN1
|
mitofusin 1 |
| chr3_-_32170850 | 4.30 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr18_+_45792035 | 4.16 |
ENSDART00000135045
|
abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr3_-_52614747 | 4.08 |
ENSDART00000154365
|
trim35-13
|
tripartite motif containing 35-13 |
| chr20_-_47732703 | 4.07 |
ENSDART00000193975
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr5_-_57635664 | 3.95 |
ENSDART00000074306
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr21_-_32487061 | 3.91 |
ENSDART00000114359
ENSDART00000131591 ENSDART00000131477 |
si:dkeyp-72g9.4
|
si:dkeyp-72g9.4 |
| chr16_+_5774977 | 3.89 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr18_-_38087875 | 3.81 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr5_-_72125551 | 3.81 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr2_-_3678029 | 3.80 |
ENSDART00000146861
|
mmp16b
|
matrix metallopeptidase 16b (membrane-inserted) |
| chr10_+_23511816 | 3.78 |
ENSDART00000127255
|
zmp:0000000924
|
zmp:0000000924 |
| chr6_-_38816500 | 3.77 |
ENSDART00000190866
ENSDART00000104124 |
cnga3a
|
cyclic nucleotide gated channel alpha 3a |
| chr1_+_33969015 | 3.74 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr14_-_27297123 | 3.68 |
ENSDART00000173423
|
pcdh11
|
protocadherin 11 |
| chr12_-_26064105 | 3.60 |
ENSDART00000168825
|
ldb3b
|
LIM domain binding 3b |
| chr1_+_7546259 | 3.56 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr21_+_12010505 | 3.56 |
ENSDART00000123522
|
aqp7
|
aquaporin 7 |
| chr21_-_131236 | 3.54 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr18_+_23193820 | 3.48 |
ENSDART00000148106
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr5_+_64732270 | 3.48 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr14_+_32837914 | 3.44 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr20_-_26001288 | 3.44 |
ENSDART00000136518
ENSDART00000063177 |
capn3b
|
calpain 3b |
| chr17_-_16069905 | 3.40 |
ENSDART00000110383
|
map7a
|
microtubule-associated protein 7a |
| chr23_+_6077503 | 3.40 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr21_+_11685009 | 3.37 |
ENSDART00000014668
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr17_+_8184649 | 3.35 |
ENSDART00000091818
|
tulp4b
|
tubby like protein 4b |
| chr12_-_26064480 | 3.32 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr21_+_11684830 | 3.30 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr5_-_20678300 | 3.30 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr25_+_31264155 | 3.29 |
ENSDART00000012256
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr4_+_9279515 | 3.29 |
ENSDART00000048707
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr11_-_33960318 | 3.22 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr3_-_49504023 | 3.21 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr6_-_49873020 | 3.21 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr17_-_12336987 | 3.20 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr7_-_7420301 | 3.17 |
ENSDART00000102620
|
six7
|
SIX homeobox 7 |
| chr19_-_7450796 | 3.13 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr16_-_32975951 | 3.11 |
ENSDART00000101969
ENSDART00000175149 |
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr19_+_15571290 | 3.09 |
ENSDART00000131134
|
foxo6b
|
forkhead box O6 b |
| chr3_+_31953145 | 3.02 |
ENSDART00000148861
|
kcnc3a
|
potassium voltage-gated channel, Shaw-related subfamily, member 3a |
| chr23_+_14097508 | 2.90 |
ENSDART00000087280
|
cacnb3a
|
calcium channel, voltage-dependent, beta 3a |
| chr18_+_7264961 | 2.89 |
ENSDART00000188461
|
CABZ01015105.1
|
|
| chr13_+_25853757 | 2.88 |
ENSDART00000132521
|
bcl11aa
|
B cell CLL/lymphoma 11Aa |
| chr10_+_41765616 | 2.87 |
ENSDART00000170682
|
rnf34b
|
ring finger protein 34b |
| chr10_-_23358357 | 2.85 |
ENSDART00000135475
|
cadm2a
|
cell adhesion molecule 2a |
| chr18_+_10820430 | 2.84 |
ENSDART00000161990
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr10_+_32561317 | 2.83 |
ENSDART00000109029
|
map6a
|
microtubule-associated protein 6a |
| chr6_+_41038757 | 2.83 |
ENSDART00000011245
|
entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr17_+_28340138 | 2.81 |
ENSDART00000033943
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr9_-_52814204 | 2.81 |
ENSDART00000140771
ENSDART00000007401 |
MAP3K13
|
si:ch211-45c16.2 |
| chr18_+_23193567 | 2.80 |
ENSDART00000190072
ENSDART00000171594 ENSDART00000181762 |
mef2aa
|
myocyte enhancer factor 2aa |
| chr15_-_14375452 | 2.79 |
ENSDART00000160675
ENSDART00000164028 ENSDART00000171642 |
dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr23_+_17981127 | 2.78 |
ENSDART00000012571
ENSDART00000145200 |
chia.6
|
chitinase, acidic.6 |
| chr23_-_27633730 | 2.77 |
ENSDART00000103639
|
arf3a
|
ADP-ribosylation factor 3a |
| chr9_-_23990416 | 2.77 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr10_+_21776911 | 2.77 |
ENSDART00000163077
ENSDART00000186093 |
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr6_+_12462079 | 2.77 |
ENSDART00000192029
ENSDART00000065385 |
nr4a2b
|
nuclear receptor subfamily 4, group A, member 2b |
| chr10_-_24371312 | 2.76 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr2_+_37110504 | 2.76 |
ENSDART00000132794
ENSDART00000042974 |
slc1a8b
|
solute carrier family 1 (glutamate transporter), member 8b |
| chr24_+_34089977 | 2.75 |
ENSDART00000157466
|
asb10
|
ankyrin repeat and SOCS box containing 10 |
| chr16_-_43356018 | 2.75 |
ENSDART00000181683
|
FO704821.1
|
|
| chr2_+_48074243 | 2.72 |
ENSDART00000056291
|
klf6b
|
Kruppel-like factor 6b |
| chr7_-_66864756 | 2.72 |
ENSDART00000184462
ENSDART00000189424 |
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr19_-_12648122 | 2.72 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr17_-_39886628 | 2.71 |
ENSDART00000002217
|
zmp:0000000545
|
zmp:0000000545 |
| chr20_+_16881883 | 2.71 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr7_+_7048245 | 2.71 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr9_-_42873700 | 2.69 |
ENSDART00000125953
|
ttn.1
|
titin, tandem duplicate 1 |
| chr16_+_25245857 | 2.68 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr1_-_8917902 | 2.60 |
ENSDART00000137900
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr25_-_23745765 | 2.58 |
ENSDART00000089278
|
si:ch211-236l14.4
|
si:ch211-236l14.4 |
| chr25_+_14087045 | 2.57 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr20_+_34455645 | 2.57 |
ENSDART00000135789
|
mettl11b
|
methyltransferase like 11B |
| chr21_-_22910520 | 2.57 |
ENSDART00000065567
ENSDART00000191792 |
guca1d
|
guanylate cyclase activator 1d |
| chr11_-_19650781 | 2.56 |
ENSDART00000165595
|
SYNPR
|
si:dkey-30j16.3 |
| chr25_+_20077225 | 2.55 |
ENSDART00000136543
|
tnni4b.1
|
troponin I4b, tandem duplicate 1 |
| chrM_-_15232 | 2.52 |
ENSDART00000093623
|
mt-nd6
|
NADH dehydrogenase 6, mitochondrial |
| chr3_-_42981739 | 2.52 |
ENSDART00000167844
|
mafk
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr23_-_19953089 | 2.49 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr23_+_36601984 | 2.41 |
ENSDART00000128598
|
igfbp6b
|
insulin-like growth factor binding protein 6b |
| chr15_-_22139566 | 2.38 |
ENSDART00000149017
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr14_-_46451249 | 2.37 |
ENSDART00000058466
|
fgfbp2a
|
fibroblast growth factor binding protein 2a |
| chr19_-_10394931 | 2.35 |
ENSDART00000191549
|
zgc:194578
|
zgc:194578 |
| chr16_-_19890303 | 2.34 |
ENSDART00000147161
ENSDART00000079159 |
hdac9b
|
histone deacetylase 9b |
| chr17_-_50233493 | 2.32 |
ENSDART00000172266
|
fosaa
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
| chr2_+_48073972 | 2.32 |
ENSDART00000186442
|
klf6b
|
Kruppel-like factor 6b |
| chr5_-_64511428 | 2.29 |
ENSDART00000016321
|
rxrab
|
retinoid x receptor, alpha b |
| chr8_+_52637507 | 2.29 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr3_+_28953274 | 2.27 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr17_-_50234004 | 2.26 |
ENSDART00000058706
|
fosaa
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
| chr1_-_22652424 | 2.22 |
ENSDART00000036797
|
uchl1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr18_-_6832337 | 2.20 |
ENSDART00000139360
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr1_+_19708508 | 2.15 |
ENSDART00000054581
ENSDART00000131206 |
march1
|
membrane-associated ring finger (C3HC4) 1 |
| chr25_+_3677650 | 2.14 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr8_-_32497815 | 2.12 |
ENSDART00000122359
|
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr14_-_2318590 | 2.06 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr15_+_28940352 | 2.03 |
ENSDART00000154085
|
gipr
|
gastric inhibitory polypeptide receptor |
| chr25_-_28384954 | 2.03 |
ENSDART00000073500
|
ptprz1a
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1a |
| chr11_+_11200550 | 2.02 |
ENSDART00000181339
ENSDART00000187116 |
myom2a
|
myomesin 2a |
| chr9_+_29643036 | 2.02 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr9_-_23891102 | 2.02 |
ENSDART00000186799
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr12_-_6905375 | 2.00 |
ENSDART00000152322
|
pcdh15b
|
protocadherin-related 15b |
| chr13_+_22480857 | 1.99 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr7_+_6969909 | 1.94 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr20_+_33532296 | 1.91 |
ENSDART00000153153
|
kcnf1a
|
potassium voltage-gated channel, subfamily F, member 1a |
| chr8_-_12264486 | 1.89 |
ENSDART00000091612
ENSDART00000135812 |
dab2ipa
|
DAB2 interacting protein a |
| chr1_+_38858399 | 1.88 |
ENSDART00000165454
|
CU915762.1
|
|
| chr9_-_43073960 | 1.87 |
ENSDART00000059460
|
ttn.2
|
titin, tandem duplicate 2 |
| chr2_-_8611675 | 1.86 |
ENSDART00000138223
|
si:ch211-71m22.1
|
si:ch211-71m22.1 |
| chr17_-_43558494 | 1.86 |
ENSDART00000103830
|
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr19_-_31035155 | 1.85 |
ENSDART00000161882
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr6_+_57541776 | 1.83 |
ENSDART00000157330
|
necab3
|
N-terminal EF-hand calcium binding protein 3 |
| chr7_+_72882446 | 1.83 |
ENSDART00000131113
ENSDART00000174223 |
ADGRL3
|
adhesion G protein-coupled receptor L3 |
| chr19_-_31035325 | 1.82 |
ENSDART00000147504
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr23_+_17980875 | 1.78 |
ENSDART00000163452
|
chia.6
|
chitinase, acidic.6 |
| chr3_+_32403758 | 1.78 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr5_+_69808763 | 1.77 |
ENSDART00000143482
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr2_-_36925561 | 1.76 |
ENSDART00000187690
|
map1sb
|
microtubule-associated protein 1Sb |
| chr20_-_2134620 | 1.76 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr16_-_16590489 | 1.71 |
ENSDART00000190021
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr7_-_42206720 | 1.70 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr21_-_40083432 | 1.69 |
ENSDART00000141160
ENSDART00000191195 |
slc13a5a
|
info solute carrier family 13 (sodium-dependent citrate transporter), member 5a |
| chr14_-_7245971 | 1.66 |
ENSDART00000108796
|
stox2b
|
storkhead box 2b |
| chr16_+_29663809 | 1.65 |
ENSDART00000191336
|
tmod4
|
tropomodulin 4 (muscle) |
| chr7_+_69841017 | 1.62 |
ENSDART00000169107
|
FO818704.1
|
|
| chr6_-_12459412 | 1.58 |
ENSDART00000090266
ENSDART00000144028 |
gpd2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr23_+_20110086 | 1.58 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr20_+_6756247 | 1.56 |
ENSDART00000167344
|
igfbp3
|
insulin-like growth factor binding protein 3 |
| chr13_+_31172833 | 1.55 |
ENSDART00000176378
|
CR931802.3
|
|
| chr16_+_20161805 | 1.54 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr10_-_10607118 | 1.48 |
ENSDART00000101089
|
dbh
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr24_-_6647275 | 1.47 |
ENSDART00000161494
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr17_-_5583345 | 1.46 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr1_-_37383741 | 1.45 |
ENSDART00000193155
ENSDART00000191887 ENSDART00000189077 |
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr17_+_43623598 | 1.42 |
ENSDART00000154138
|
znf365
|
zinc finger protein 365 |
| chr11_-_26701611 | 1.41 |
ENSDART00000083010
|
acad9
|
acyl-CoA dehydrogenase family, member 9 |
| chr16_+_32029090 | 1.40 |
ENSDART00000041054
|
tmc4
|
transmembrane channel-like 4 |
| chr12_-_15584479 | 1.40 |
ENSDART00000150134
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr21_+_43559123 | 1.40 |
ENSDART00000151212
|
gpr185a
|
G protein-coupled receptor 185 a |
| chr2_-_15324837 | 1.39 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr11_-_40030139 | 1.39 |
ENSDART00000021916
|
uts2b
|
urotensin 2, beta |
| chr22_-_13649588 | 1.38 |
ENSDART00000131877
|
si:ch211-279g13.1
|
si:ch211-279g13.1 |
| chr5_+_19094153 | 1.36 |
ENSDART00000186525
ENSDART00000064752 |
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr22_+_38301365 | 1.35 |
ENSDART00000137339
|
traf5
|
Tnf receptor-associated factor 5 |
| chr12_+_8003872 | 1.34 |
ENSDART00000180300
ENSDART00000126661 |
rhobtb1
|
Rho-related BTB domain containing 1 |
| chr21_-_5799122 | 1.34 |
ENSDART00000129351
ENSDART00000151202 |
ccni
|
cyclin I |
| chr18_+_39327010 | 1.33 |
ENSDART00000012164
|
tmod2
|
tropomodulin 2 |
| chr8_+_30456161 | 1.32 |
ENSDART00000085894
|
pgm5
|
phosphoglucomutase 5 |
| chr1_-_11075403 | 1.31 |
ENSDART00000102903
ENSDART00000170290 |
dmd
|
dystrophin |
| chr7_+_42206847 | 1.28 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr10_-_41450367 | 1.27 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr6_-_19351495 | 1.26 |
ENSDART00000164287
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr21_+_40944530 | 1.26 |
ENSDART00000022976
|
kctd16b
|
potassium channel tetramerization domain containing 16b |
| chr19_+_24394560 | 1.23 |
ENSDART00000142506
|
si:dkey-81h8.1
|
si:dkey-81h8.1 |
| chr19_+_2590182 | 1.22 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr16_-_17541890 | 1.20 |
ENSDART00000131328
|
clcn1b
|
chloride channel, voltage-sensitive 1b |
| chr7_+_39006837 | 1.19 |
ENSDART00000173735
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr8_-_32497581 | 1.19 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr7_+_40093662 | 1.19 |
ENSDART00000053011
|
si:ch73-174h16.4
|
si:ch73-174h16.4 |
| chr2_+_8779164 | 1.19 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr7_-_31321027 | 1.18 |
ENSDART00000186878
|
CR356242.1
|
|
| chr8_-_42594380 | 1.15 |
ENSDART00000140126
ENSDART00000135238 ENSDART00000192764 |
dok2
|
docking protein 2 |
| chr3_+_32933663 | 1.15 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr17_-_28770800 | 1.11 |
ENSDART00000156485
|
opn6b
|
opsin 6, group member b |
| chr25_+_10793478 | 1.11 |
ENSDART00000058339
ENSDART00000134923 |
ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr8_-_42238543 | 1.10 |
ENSDART00000062697
|
gfra2a
|
GDNF family receptor alpha 2a |
| chr5_+_19094462 | 1.09 |
ENSDART00000190596
|
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr24_+_38155830 | 1.08 |
ENSDART00000152019
|
si:ch211-234p6.5
|
si:ch211-234p6.5 |
| chr14_+_11430796 | 1.07 |
ENSDART00000165275
|
si:ch211-153b23.3
|
si:ch211-153b23.3 |
| chr14_+_23811808 | 1.03 |
ENSDART00000014411
|
kctd16a
|
potassium channel tetramerization domain containing 16a |
| chr14_+_30910114 | 1.01 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr8_-_4972916 | 1.00 |
ENSDART00000101576
|
tmem230b
|
transmembrane protein 230b |
| chr24_+_20960216 | 1.00 |
ENSDART00000133008
|
si:ch211-161h7.8
|
si:ch211-161h7.8 |
| chr6_-_37468971 | 0.99 |
ENSDART00000126379
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mef2aa+mef2ab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 1.3 | 7.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.1 | 3.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 1.1 | 15.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 1.0 | 5.0 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.9 | 2.7 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.8 | 4.0 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.7 | 2.9 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.7 | 9.2 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.6 | 3.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.6 | 4.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.6 | 2.4 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.5 | 2.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.5 | 1.5 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.5 | 1.0 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.4 | 6.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.4 | 3.8 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.4 | 4.6 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.4 | 4.6 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.4 | 1.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.4 | 2.9 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 | 2.4 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.4 | 3.5 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.4 | 3.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.4 | 4.0 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.4 | 3.2 | GO:0071331 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.3 | 5.0 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.3 | 9.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.3 | 4.8 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.3 | 3.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.3 | 2.8 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.3 | 3.4 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.3 | 5.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.3 | 1.8 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 4.5 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.2 | 2.4 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.2 | 1.9 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 3.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 6.8 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.2 | 3.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.2 | 2.3 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.2 | 0.9 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.2 | 7.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 1.7 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.2 | 0.8 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 2.8 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.2 | 0.5 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 | 1.0 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 32.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 1.9 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 2.6 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 1.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.7 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 8.4 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.1 | 1.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 1.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 1.3 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 3.5 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 0.4 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 3.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 3.9 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 2.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.0 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 2.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 1.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.6 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 1.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 0.4 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 0.4 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 0.7 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 0.5 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 1.7 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 1.8 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 5.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 2.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 8.5 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.1 | 1.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.6 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 5.9 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 0.2 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.1 | 1.9 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.3 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.1 | 1.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 5.5 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.7 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 2.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 2.8 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.2 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 1.0 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 4.2 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.3 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.3 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 2.3 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 2.8 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 2.5 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.1 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 1.4 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 0.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.8 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.6 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.5 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.6 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.7 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.6 | GO:0061245 | establishment or maintenance of apical/basal cell polarity(GO:0035088) establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) establishment or maintenance of bipolar cell polarity(GO:0061245) |
| 0.0 | 0.6 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.2 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 2.1 | GO:0033674 | positive regulation of kinase activity(GO:0033674) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.3 | 8.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 3.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.3 | 5.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 5.8 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 11.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.9 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 5.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 3.0 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.2 | 8.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 2.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 3.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 2.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 5.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 4.7 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 5.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 2.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 2.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 5.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 2.6 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 3.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 1.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 3.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 4.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 3.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 3.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 1.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.8 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 9.6 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 4.4 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 17.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 5.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 2.7 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 2.6 | 7.7 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 2.3 | 6.8 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 1.4 | 15.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.2 | 4.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.8 | 3.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.7 | 9.2 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.6 | 3.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.6 | 3.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.6 | 4.8 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.5 | 1.5 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.5 | 1.4 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.5 | 1.4 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.4 | 8.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.4 | 3.6 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.4 | 1.7 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.4 | 4.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.4 | 2.0 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.4 | 15.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.4 | 2.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.4 | 4.0 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 27.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 7.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 2.6 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.3 | 5.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 2.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.3 | 3.8 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.3 | 2.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 2.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 0.7 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 3.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.2 | 3.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 0.8 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.2 | 0.7 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.2 | 2.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 2.8 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 1.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 0.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.9 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.1 | 1.0 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 3.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 2.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 6.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 3.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 3.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 3.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 3.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 3.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.3 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 1.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 1.0 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 2.1 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 6.0 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.1 | 1.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 3.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 1.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 3.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 2.8 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.0 | 1.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 1.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.9 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 1.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 13.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.7 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 2.2 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 23.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 1.4 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 1.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 3.0 | GO:0043492 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 4.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 1.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.5 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 3.2 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 6.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 2.8 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 2.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 5.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 1.0 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.3 | GO:0005516 | calmodulin binding(GO:0005516) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 1.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 6.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 1.2 | 12.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 1.2 | 3.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.7 | 6.7 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.4 | 0.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.4 | 9.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.3 | 4.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.3 | 6.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 6.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 5.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 1.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 1.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 6.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 6.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.0 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 2.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.6 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |