Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for mecp2
Z-value: 1.70
Transcription factors associated with mecp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mecp2
|
ENSDARG00000014218 | methyl CpG binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mecp2 | dr11_v1_chr8_-_7637626_7637640 | 0.62 | 4.5e-03 | Click! |
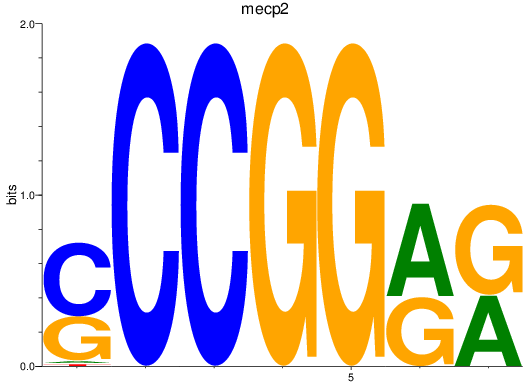
Activity profile of mecp2 motif
Sorted Z-values of mecp2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_68563862 | 8.09 |
ENSDART00000182970
|
BX548011.2
|
|
| chr4_-_77557279 | 7.92 |
ENSDART00000180113
|
AL935186.10
|
|
| chr4_-_68564988 | 7.11 |
ENSDART00000191212
|
BX548011.3
|
|
| chr5_-_825920 | 5.24 |
ENSDART00000126982
|
zgc:158463
|
zgc:158463 |
| chr3_+_31396149 | 2.98 |
ENSDART00000151423
ENSDART00000193580 |
c1ql3b
|
complement component 1, q subcomponent-like 3b |
| chr11_+_39969048 | 2.75 |
ENSDART00000193693
|
per3
|
period circadian clock 3 |
| chr19_-_11846958 | 2.66 |
ENSDART00000148516
|
ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr19_-_47587719 | 2.58 |
ENSDART00000111108
|
CABZ01071972.1
|
|
| chr9_-_32158288 | 2.55 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr4_-_77561679 | 2.53 |
ENSDART00000180809
|
AL935186.9
|
|
| chr23_+_40908583 | 2.46 |
ENSDART00000180933
|
LO017845.1
|
|
| chr5_-_13685047 | 2.38 |
ENSDART00000018351
|
zgc:65851
|
zgc:65851 |
| chr4_+_55758103 | 2.27 |
ENSDART00000185964
|
CT583728.23
|
|
| chr10_-_36633882 | 2.27 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr20_+_32523576 | 2.22 |
ENSDART00000147319
|
scml4
|
Scm polycomb group protein like 4 |
| chr19_-_6988837 | 2.21 |
ENSDART00000145741
ENSDART00000167640 |
znf384l
|
zinc finger protein 384 like |
| chr20_-_45661049 | 2.16 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr11_-_37589293 | 2.09 |
ENSDART00000172989
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr10_-_9115383 | 2.06 |
ENSDART00000139324
|
si:dkeyp-41f9.3
|
si:dkeyp-41f9.3 |
| chr19_-_677713 | 2.06 |
ENSDART00000025146
|
slc6a19a.1
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 1 |
| chr3_+_22059066 | 2.06 |
ENSDART00000155739
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr19_+_232536 | 2.04 |
ENSDART00000137880
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr15_-_18138607 | 2.03 |
ENSDART00000176690
|
CR385077.1
|
|
| chr10_-_2682198 | 1.99 |
ENSDART00000183727
|
pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr16_+_34528409 | 1.96 |
ENSDART00000144718
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr9_+_56422311 | 1.90 |
ENSDART00000171958
|
gpr39
|
G protein-coupled receptor 39 |
| chr13_+_51853716 | 1.89 |
ENSDART00000175341
ENSDART00000187855 |
LT631684.1
|
|
| chr8_-_4327473 | 1.89 |
ENSDART00000134378
|
cux2b
|
cut-like homeobox 2b |
| chr6_+_41554794 | 1.88 |
ENSDART00000165424
|
srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr8_+_48966165 | 1.86 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr20_-_52939501 | 1.85 |
ENSDART00000166508
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr2_-_50372467 | 1.85 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein like 2b |
| chr17_-_6759006 | 1.82 |
ENSDART00000184692
ENSDART00000180530 |
vsnl1b
|
visinin-like 1b |
| chr4_+_3438510 | 1.77 |
ENSDART00000155320
|
atxn7l1
|
ataxin 7-like 1 |
| chr14_-_2355833 | 1.75 |
ENSDART00000157677
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.6 |
| chr4_+_23223881 | 1.73 |
ENSDART00000133056
ENSDART00000089126 |
trhde.1
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 1 |
| chr12_+_22680115 | 1.73 |
ENSDART00000152879
|
ablim2
|
actin binding LIM protein family, member 2 |
| chr21_-_10446405 | 1.72 |
ENSDART00000167948
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr4_-_1360495 | 1.69 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr12_+_33038757 | 1.68 |
ENSDART00000153146
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr10_+_41668483 | 1.66 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr1_-_54706039 | 1.66 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr8_+_48965767 | 1.63 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr17_+_389218 | 1.62 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr15_-_30816370 | 1.57 |
ENSDART00000142982
ENSDART00000050649 ENSDART00000136901 ENSDART00000100194 |
msi2b
|
musashi RNA-binding protein 2b |
| chr19_+_41479990 | 1.57 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr3_+_14463941 | 1.56 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr3_-_27880229 | 1.55 |
ENSDART00000151404
|
abat
|
4-aminobutyrate aminotransferase |
| chr21_-_8513192 | 1.54 |
ENSDART00000084378
|
crb2a
|
crumbs family member 2a |
| chr21_-_2814709 | 1.54 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr14_-_2050057 | 1.51 |
ENSDART00000112875
|
PCDHB15
|
protocadherin beta 15 |
| chr15_-_37921998 | 1.48 |
ENSDART00000193597
ENSDART00000181443 ENSDART00000168790 |
si:dkey-238d18.5
|
si:dkey-238d18.5 |
| chr25_-_35145920 | 1.46 |
ENSDART00000157137
|
si:dkey-261m9.19
|
si:dkey-261m9.19 |
| chr9_+_17787864 | 1.45 |
ENSDART00000013111
|
dgkh
|
diacylglycerol kinase, eta |
| chr19_-_9503473 | 1.45 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr6_-_21988375 | 1.44 |
ENSDART00000161257
|
plxnb1b
|
plexin b1b |
| chr15_+_2559875 | 1.44 |
ENSDART00000178505
|
SH2B2
|
SH2B adaptor protein 2 |
| chr17_+_52300018 | 1.43 |
ENSDART00000190302
|
esrrb
|
estrogen-related receptor beta |
| chr9_-_55790458 | 1.43 |
ENSDART00000161247
|
sowahca
|
sosondowah ankyrin repeat domain family Ca |
| chr17_-_11418513 | 1.42 |
ENSDART00000064412
ENSDART00000151847 |
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr25_+_8063455 | 1.41 |
ENSDART00000073919
|
kcnc1b
|
potassium voltage-gated channel, Shaw-related subfamily, member 1b |
| chr6_+_57741039 | 1.41 |
ENSDART00000169175
|
SNTA1
|
syntrophin alpha 1 |
| chr10_-_44981295 | 1.41 |
ENSDART00000166528
|
purbb
|
purine-rich element binding protein Bb |
| chr15_+_28685625 | 1.41 |
ENSDART00000188797
ENSDART00000166036 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr20_-_24122881 | 1.39 |
ENSDART00000131857
|
bach2b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2b |
| chr19_+_48372446 | 1.39 |
ENSDART00000161466
|
abhd5b
|
abhydrolase domain containing 5b |
| chr20_+_4060839 | 1.39 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr13_-_31166544 | 1.38 |
ENSDART00000146250
ENSDART00000132129 ENSDART00000139591 |
mapk8a
|
mitogen-activated protein kinase 8a |
| chr23_+_2361184 | 1.38 |
ENSDART00000184469
|
CABZ01048666.1
|
|
| chr6_+_58406014 | 1.37 |
ENSDART00000044241
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr4_-_12102025 | 1.36 |
ENSDART00000048391
ENSDART00000023894 |
braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr4_+_20263097 | 1.35 |
ENSDART00000138820
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr5_+_22633188 | 1.33 |
ENSDART00000128781
|
mtnr1c
|
melatonin receptor 1C |
| chr9_-_135774 | 1.33 |
ENSDART00000160435
|
FQ377903.1
|
|
| chr22_+_15331214 | 1.33 |
ENSDART00000136566
|
sult3st4
|
sulfotransferase family 3, cytosolic sulfotransferase 4 |
| chr18_-_46881108 | 1.32 |
ENSDART00000190084
|
gramd1bb
|
GRAM domain containing 1Bb |
| chr15_+_30158652 | 1.31 |
ENSDART00000190682
|
nlk2
|
nemo-like kinase, type 2 |
| chr8_+_2757821 | 1.30 |
ENSDART00000051403
ENSDART00000160551 |
sh3glb2a
|
SH3-domain GRB2-like endophilin B2a |
| chr19_+_3056450 | 1.30 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr3_+_48842918 | 1.29 |
ENSDART00000159420
|
hs3st3b1a
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1a |
| chr11_+_19781053 | 1.29 |
ENSDART00000029680
ENSDART00000182743 |
cadpsa
|
Ca2+-dependent activator protein for secretion a |
| chr21_-_42100471 | 1.28 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr3_-_25054002 | 1.28 |
ENSDART00000086768
|
ep300b
|
E1A binding protein p300 b |
| chr16_-_13388821 | 1.28 |
ENSDART00000144062
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr23_-_44677315 | 1.20 |
ENSDART00000143054
|
kif1c
|
kinesin family member 1C |
| chr8_-_3773374 | 1.18 |
ENSDART00000115036
|
bicdl1
|
BICD family like cargo adaptor 1 |
| chr20_+_52389858 | 1.18 |
ENSDART00000185863
ENSDART00000166651 |
arhgap39
|
Rho GTPase activating protein 39 |
| chr18_-_1143996 | 1.17 |
ENSDART00000189186
ENSDART00000136140 |
hcn4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| chr7_+_1534820 | 1.16 |
ENSDART00000192997
|
tox4b
|
TOX high mobility group box family member 4 b |
| chr11_-_38510469 | 1.16 |
ENSDART00000162305
|
si:ch211-117k10.3
|
si:ch211-117k10.3 |
| chr17_+_43629008 | 1.15 |
ENSDART00000184185
ENSDART00000181681 |
znf365
|
zinc finger protein 365 |
| chr10_-_33156789 | 1.15 |
ENSDART00000192268
ENSDART00000182065 ENSDART00000081170 |
cux1a
|
cut-like homeobox 1a |
| chr23_-_27694960 | 1.15 |
ENSDART00000135129
|
IKZF4
|
si:dkey-166n8.9 |
| chr20_+_474288 | 1.14 |
ENSDART00000026794
|
nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr16_-_52966811 | 1.14 |
ENSDART00000153758
ENSDART00000009256 |
brd9
|
bromodomain containing 9 |
| chr21_+_45626136 | 1.11 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr9_+_56422517 | 1.11 |
ENSDART00000168620
|
gpr39
|
G protein-coupled receptor 39 |
| chr5_+_69950882 | 1.11 |
ENSDART00000097359
|
dnajc25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr8_+_29963024 | 1.10 |
ENSDART00000149372
|
ptch1
|
patched 1 |
| chr14_+_35024521 | 1.09 |
ENSDART00000158634
ENSDART00000170631 |
ebf3a
|
early B cell factor 3a |
| chr21_+_10021823 | 1.08 |
ENSDART00000163995
|
herc7
|
hect domain and RLD 7 |
| chr18_+_29403017 | 1.08 |
ENSDART00000176966
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr5_-_24712405 | 1.07 |
ENSDART00000033630
|
si:ch211-106a19.1
|
si:ch211-106a19.1 |
| chr15_+_44093286 | 1.06 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr14_-_2196267 | 1.06 |
ENSDART00000161674
ENSDART00000125674 |
pcdh2ab8
pcdh2ab9
|
protocadherin 2 alpha b 8 protocadherin 2 alpha b 9 |
| chr17_-_27235797 | 1.05 |
ENSDART00000130080
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr1_+_52632856 | 1.05 |
ENSDART00000011725
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr6_-_59271270 | 1.04 |
ENSDART00000126870
|
ndufa4l2b
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2b |
| chr1_-_18446161 | 1.04 |
ENSDART00000089442
|
klhl5
|
kelch-like family member 5 |
| chr15_+_45595385 | 1.04 |
ENSDART00000161937
ENSDART00000170214 ENSDART00000157450 |
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr5_-_37247654 | 1.04 |
ENSDART00000112312
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr22_-_38607504 | 1.04 |
ENSDART00000164609
|
si:ch211-126j24.1
|
si:ch211-126j24.1 |
| chr2_-_2642476 | 1.03 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr9_+_27411502 | 1.02 |
ENSDART00000143994
|
si:dkey-193n17.9
|
si:dkey-193n17.9 |
| chr6_-_607063 | 1.01 |
ENSDART00000189900
|
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr18_+_50961953 | 1.00 |
ENSDART00000158768
|
ppfia1
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
| chr15_+_45994123 | 1.00 |
ENSDART00000124704
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr13_-_301309 | 1.00 |
ENSDART00000131747
|
chs1
|
chitin synthase 1 |
| chr3_+_17806213 | 0.99 |
ENSDART00000055890
|
znf385c
|
zinc finger protein 385C |
| chr5_+_45140914 | 0.99 |
ENSDART00000172702
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr9_+_30585552 | 0.99 |
ENSDART00000160590
ENSDART00000189815 ENSDART00000143766 |
tbc1d4
|
TBC1 domain family, member 4 |
| chr9_+_24159725 | 0.98 |
ENSDART00000137756
|
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr20_+_715739 | 0.97 |
ENSDART00000136768
|
myo6a
|
myosin VIa |
| chr25_+_3994823 | 0.97 |
ENSDART00000154020
|
eps8l2
|
EPS8 like 2 |
| chr21_-_45891262 | 0.95 |
ENSDART00000169816
|
galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr15_+_31471808 | 0.95 |
ENSDART00000110078
|
or102-3
|
odorant receptor, family C, subfamily 102, member 3 |
| chr6_+_58915889 | 0.94 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr21_+_8427059 | 0.94 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr6_-_36795111 | 0.94 |
ENSDART00000160669
ENSDART00000104256 ENSDART00000187751 ENSDART00000161928 ENSDART00000183264 |
opa1
|
optic atrophy 1 (autosomal dominant) |
| chr3_+_13637383 | 0.94 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr18_+_28564640 | 0.94 |
ENSDART00000016983
|
spon1a
|
spondin 1a |
| chr4_+_121709 | 0.94 |
ENSDART00000186461
ENSDART00000172255 |
crebl2
|
cAMP responsive element binding protein-like 2 |
| chr7_+_1442059 | 0.93 |
ENSDART00000173391
|
si:cabz01090193.1
|
si:cabz01090193.1 |
| chr5_+_15992655 | 0.90 |
ENSDART00000182148
|
znrf3
|
zinc and ring finger 3 |
| chr1_-_23370395 | 0.90 |
ENSDART00000143014
ENSDART00000126785 ENSDART00000159138 |
pds5a
|
PDS5 cohesin associated factor A |
| chr18_+_507835 | 0.89 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr18_+_45228691 | 0.88 |
ENSDART00000127953
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr11_+_45436703 | 0.88 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr5_-_19861766 | 0.88 |
ENSDART00000088904
|
gltpa
|
glycolipid transfer protein a |
| chr9_-_52206336 | 0.88 |
ENSDART00000114222
|
tanc1b
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1b |
| chr24_+_792429 | 0.88 |
ENSDART00000082523
|
impa2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr25_+_4581214 | 0.88 |
ENSDART00000185552
|
CABZ01068600.1
|
|
| chr5_-_29488245 | 0.87 |
ENSDART00000047719
ENSDART00000141154 ENSDART00000171165 |
cacna1ba
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, a |
| chr18_-_16590056 | 0.87 |
ENSDART00000143744
|
mgat4c
|
mgat4 family, member C |
| chr24_-_33276139 | 0.87 |
ENSDART00000128943
|
nrbp2b
|
nuclear receptor binding protein 2b |
| chr13_-_40120252 | 0.87 |
ENSDART00000157852
|
crtac1b
|
cartilage acidic protein 1b |
| chr3_+_58655375 | 0.86 |
ENSDART00000186908
|
FP016018.4
|
|
| chr15_-_47895200 | 0.85 |
ENSDART00000027060
|
DMWD
|
zmp:0000000529 |
| chr13_+_681628 | 0.84 |
ENSDART00000016604
|
abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr16_+_68069 | 0.84 |
ENSDART00000185385
ENSDART00000159652 |
sox4b
|
SRY (sex determining region Y)-box 4b |
| chr6_-_57722816 | 0.83 |
ENSDART00000186163
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr6_+_39222598 | 0.83 |
ENSDART00000154991
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr8_+_29962635 | 0.83 |
ENSDART00000007640
|
ptch1
|
patched 1 |
| chr20_+_52458765 | 0.83 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr12_+_28749189 | 0.82 |
ENSDART00000013980
|
tbx21
|
T-box 21 |
| chr2_+_39108339 | 0.82 |
ENSDART00000085675
|
clstn2
|
calsyntenin 2 |
| chr3_+_35005062 | 0.81 |
ENSDART00000181163
|
prkcbb
|
protein kinase C, beta b |
| chr12_-_18961289 | 0.81 |
ENSDART00000168405
|
ep300a
|
E1A binding protein p300 a |
| chr17_+_22587356 | 0.81 |
ENSDART00000157328
|
birc6
|
baculoviral IAP repeat containing 6 |
| chr12_-_3705862 | 0.81 |
ENSDART00000193864
ENSDART00000185857 |
CABZ01069040.1
|
|
| chr8_-_22739757 | 0.81 |
ENSDART00000182167
ENSDART00000171891 |
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr2_-_5399437 | 0.81 |
ENSDART00000132411
|
si:ch1073-184j22.2
|
si:ch1073-184j22.2 |
| chr10_+_44042033 | 0.81 |
ENSDART00000190006
ENSDART00000046172 |
cryba4
|
crystallin, beta A4 |
| chr10_-_41156348 | 0.80 |
ENSDART00000058622
|
aak1b
|
AP2 associated kinase 1b |
| chr10_-_22845485 | 0.80 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr7_-_33921366 | 0.80 |
ENSDART00000052397
|
pias1a
|
protein inhibitor of activated STAT, 1a |
| chr13_-_36525982 | 0.77 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr19_+_9032073 | 0.77 |
ENSDART00000127755
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr6_-_59271436 | 0.77 |
ENSDART00000174588
|
ndufa4l2b
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2b |
| chr12_+_25845494 | 0.76 |
ENSDART00000170582
|
glud1b
|
glutamate dehydrogenase 1b |
| chr2_-_32501501 | 0.76 |
ENSDART00000181309
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr22_-_13544244 | 0.76 |
ENSDART00000110136
|
cntnap5b
|
contactin associated protein-like 5b |
| chr16_-_2807527 | 0.76 |
ENSDART00000148927
|
tgfb2
|
transforming growth factor, beta 2 |
| chr5_-_49012569 | 0.75 |
ENSDART00000184690
|
si:dkey-172m14.2
|
si:dkey-172m14.2 |
| chr18_-_399554 | 0.75 |
ENSDART00000164374
ENSDART00000186311 ENSDART00000181816 ENSDART00000181892 |
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr10_-_43392667 | 0.75 |
ENSDART00000183033
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr15_+_23208042 | 0.75 |
ENSDART00000006085
|
cbl
|
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr5_+_22791686 | 0.74 |
ENSDART00000014806
|
npas2
|
neuronal PAS domain protein 2 |
| chr1_+_38858399 | 0.74 |
ENSDART00000165454
|
CU915762.1
|
|
| chr1_-_53407448 | 0.74 |
ENSDART00000160033
ENSDART00000172322 |
elmod2
|
ELMO/CED-12 domain containing 2 |
| chr1_-_23557877 | 0.73 |
ENSDART00000145942
|
fam184b
|
family with sequence similarity 184, member B |
| chr2_+_24507597 | 0.72 |
ENSDART00000133109
|
rps28
|
ribosomal protein S28 |
| chr9_+_1505206 | 0.72 |
ENSDART00000093427
ENSDART00000137230 |
pde11a
|
phosphodiesterase 11a |
| chr18_+_50461981 | 0.72 |
ENSDART00000158761
|
CU896640.1
|
|
| chr10_+_34377697 | 0.71 |
ENSDART00000189441
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr11_+_2687395 | 0.70 |
ENSDART00000082510
|
b3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 |
| chr21_-_13751535 | 0.69 |
ENSDART00000111666
|
npdc1a
|
neural proliferation, differentiation and control, 1a |
| chr7_+_9922607 | 0.67 |
ENSDART00000184532
ENSDART00000113396 |
cers3a
|
ceramide synthase 3a |
| chr13_-_51922290 | 0.66 |
ENSDART00000168648
|
srfb
|
serum response factor b |
| chr5_+_26079178 | 0.66 |
ENSDART00000145920
|
si:dkey-201c13.2
|
si:dkey-201c13.2 |
| chr10_+_39476600 | 0.65 |
ENSDART00000135756
|
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr1_+_31864404 | 0.64 |
ENSDART00000075260
|
inab
|
internexin neuronal intermediate filament protein, alpha b |
| chr3_-_39627412 | 0.64 |
ENSDART00000123292
|
si:dkey-27o4.1
|
si:dkey-27o4.1 |
| chr8_+_50190742 | 0.64 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr24_-_20808283 | 0.64 |
ENSDART00000143759
|
vipr1b
|
vasoactive intestinal peptide receptor 1b |
| chr25_+_35067318 | 0.64 |
ENSDART00000186828
|
CU302436.1
|
|
| chr19_+_43822968 | 0.64 |
ENSDART00000171041
|
eloa
|
elongin A |
| chr13_+_14006118 | 0.63 |
ENSDART00000131875
ENSDART00000089528 |
atrn
|
attractin |
| chr17_-_5860222 | 0.62 |
ENSDART00000058894
|
si:ch73-340m8.2
|
si:ch73-340m8.2 |
| chr4_-_789645 | 0.61 |
ENSDART00000164441
|
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
Network of associatons between targets according to the STRING database.
First level regulatory network of mecp2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.5 | 2.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.5 | 2.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.4 | 1.8 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.3 | 1.4 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.3 | 1.6 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.3 | 1.6 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.3 | 0.9 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.3 | 1.2 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.3 | 1.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.3 | 4.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 1.0 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 0.9 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 0.9 | GO:0046324 | positive regulation of fat cell differentiation(GO:0045600) regulation of glucose import(GO:0046324) |
| 0.2 | 1.8 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.2 | 0.9 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.2 | 1.9 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.2 | 2.1 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.2 | 1.0 | GO:0019079 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 1.0 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.2 | 0.6 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.2 | 0.6 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 0.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 0.5 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.2 | 1.0 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 1.7 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.2 | 0.5 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.1 | 0.7 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.1 | 0.4 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 2.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.0 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.8 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.8 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 | 1.7 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 0.9 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 1.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 1.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.5 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.1 | 1.0 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.1 | 0.4 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.5 | GO:0007618 | mating(GO:0007618) |
| 0.1 | 0.6 | GO:1901490 | protein kinase D signaling(GO:0089700) regulation of lymphangiogenesis(GO:1901490) |
| 0.1 | 0.6 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.3 | GO:0048660 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 0.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 1.8 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.6 | GO:0071623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 2.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.8 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.8 | GO:0072676 | lymphocyte migration(GO:0072676) |
| 0.1 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.5 | GO:0090075 | relaxation of muscle(GO:0090075) |
| 0.1 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.2 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.1 | 0.5 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.1 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.7 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 0.2 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 1.4 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 0.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.2 | GO:0071867 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 0.7 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.1 | 1.0 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 1.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 1.0 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.5 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0007613 | memory(GO:0007613) |
| 0.0 | 0.2 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.4 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.9 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.4 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 1.2 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.7 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 2.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.5 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.7 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.3 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.5 | GO:0061647 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.8 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 1.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.6 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.5 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 3.6 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 0.6 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0032728 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.8 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 1.1 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 1.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 5.4 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 1.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 1.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.5 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.5 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.5 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.0 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 4.1 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 2.1 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 1.1 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 3.3 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.6 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.3 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.1 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.0 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.6 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0031213 | RSF complex(GO:0031213) |
| 0.5 | 2.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.3 | 1.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.3 | 1.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.3 | 1.6 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 0.7 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.2 | 1.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 2.1 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.2 | 1.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.7 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 2.1 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 1.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.0 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.8 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 1.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 1.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.6 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 0.5 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 1.7 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.8 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 2.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.0 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 3.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.4 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 1.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 2.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 0.3 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 1.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.9 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.5 | 1.9 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.4 | 1.6 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.4 | 1.2 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.4 | 2.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.4 | 4.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 2.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.3 | 0.9 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 2.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 2.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.3 | 0.9 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 0.7 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.2 | 1.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 1.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 0.6 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 0.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 3.0 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 0.5 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) |
| 0.1 | 1.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.8 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 1.0 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 0.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.8 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 2.1 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.6 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 1.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.4 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 1.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 1.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 1.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.5 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.1 | 3.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.2 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.9 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 3.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.8 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.2 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.1 | 1.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.7 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 1.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 2.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.8 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 1.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 1.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 5.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.4 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.8 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.0 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 1.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 6.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.3 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 3.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 0.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 3.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.2 | 1.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.7 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 1.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.5 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.1 | 1.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 2.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |