Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
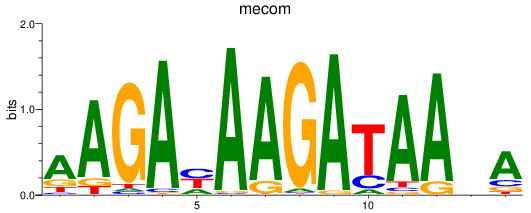
Results for mecom
Z-value: 1.11
Transcription factors associated with mecom
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mecom
|
ENSDARG00000060808 | MDS1 and EVI1 complex locus |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mecom | dr11_v1_chr15_-_35406564_35406564 | -0.82 | 1.4e-05 | Click! |
Activity profile of mecom motif
Sorted Z-values of mecom motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_26304386 | 3.59 |
ENSDART00000175416
|
otos
|
otospiralin |
| chr3_-_61203203 | 2.99 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr2_-_51794472 | 2.80 |
ENSDART00000186652
|
BX908782.3
|
|
| chr10_-_22854758 | 2.74 |
ENSDART00000079449
|
and3
|
actinodin3 |
| chr23_+_40410644 | 2.28 |
ENSDART00000056328
|
elovl4b
|
ELOVL fatty acid elongase 4b |
| chr13_+_3252950 | 2.08 |
ENSDART00000020671
|
prph2b
|
peripherin 2b (retinal degeneration, slow) |
| chr25_-_12923482 | 1.92 |
ENSDART00000161754
|
CR450808.1
|
|
| chr14_+_32838110 | 1.79 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr17_-_125091 | 1.68 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr3_-_22228602 | 1.62 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr14_+_35691889 | 1.52 |
ENSDART00000074685
|
glrbb
|
glycine receptor, beta b |
| chr10_-_8046764 | 1.48 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr15_+_22311803 | 1.47 |
ENSDART00000150182
|
hepacama
|
hepatic and glial cell adhesion molecule a |
| chr10_-_8053385 | 1.46 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr9_+_13714379 | 1.40 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr14_+_32837914 | 1.39 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr10_-_8053753 | 1.26 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr8_-_14121634 | 1.22 |
ENSDART00000184946
|
bgna
|
biglycan a |
| chr22_-_15385442 | 1.22 |
ENSDART00000090975
|
tmem264
|
transmembrane protein 264 |
| chr8_+_34731982 | 1.21 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr21_+_7582036 | 1.21 |
ENSDART00000135485
ENSDART00000027268 |
otpa
|
orthopedia homeobox a |
| chr24_-_15648636 | 1.16 |
ENSDART00000136200
|
cbln2b
|
cerebellin 2b precursor |
| chr7_-_38340674 | 1.12 |
ENSDART00000075782
|
slc7a10a
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a |
| chr6_-_46861676 | 1.06 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr7_-_8374950 | 1.06 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr10_-_31782616 | 1.05 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr10_+_11309814 | 1.04 |
ENSDART00000145673
|
si:ch211-126i22.5
|
si:ch211-126i22.5 |
| chr11_-_13126505 | 1.03 |
ENSDART00000158377
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr6_-_40697585 | 1.02 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr14_-_3070613 | 1.01 |
ENSDART00000193729
ENSDART00000090196 |
slc35a4
|
solute carrier family 35, member A4 |
| chr8_-_31369161 | 1.00 |
ENSDART00000019937
|
gadd45ga
|
growth arrest and DNA-damage-inducible, gamma a |
| chr8_+_20880848 | 0.98 |
ENSDART00000134488
ENSDART00000138605 ENSDART00000192234 |
si:ch73-196i15.3
|
si:ch73-196i15.3 |
| chr1_+_31110817 | 0.95 |
ENSDART00000137863
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr6_-_50203682 | 0.94 |
ENSDART00000083999
ENSDART00000143050 |
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr6_-_10963887 | 0.92 |
ENSDART00000151102
|
notum2
|
notum pectinacetylesterase 2 |
| chr14_-_6527010 | 0.91 |
ENSDART00000125058
|
nipsnap3a
|
nipsnap homolog 3A (C. elegans) |
| chr25_-_12935065 | 0.90 |
ENSDART00000167362
|
ccl39.7
|
chemokine (C-C motif) ligand 39, duplicate 7 |
| chr18_+_8346920 | 0.89 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr18_-_17485419 | 0.86 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr21_+_15704556 | 0.86 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr3_-_32817274 | 0.84 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr15_+_7054754 | 0.83 |
ENSDART00000149800
|
foxl2a
|
forkhead box L2a |
| chr5_-_10946232 | 0.81 |
ENSDART00000163139
ENSDART00000031265 |
rtn4r
|
reticulon 4 receptor |
| chr22_+_10752511 | 0.80 |
ENSDART00000081188
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr10_-_30785501 | 0.79 |
ENSDART00000020054
|
opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr20_-_34801181 | 0.78 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr14_+_36738069 | 0.76 |
ENSDART00000105590
|
tdo2a
|
tryptophan 2,3-dioxygenase a |
| chr18_+_16330025 | 0.75 |
ENSDART00000142353
|
nts
|
neurotensin |
| chr22_+_10752787 | 0.73 |
ENSDART00000186542
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr6_-_29195642 | 0.73 |
ENSDART00000078625
|
dpt
|
dermatopontin |
| chr4_-_16353733 | 0.72 |
ENSDART00000186785
|
lum
|
lumican |
| chr4_+_41227127 | 0.72 |
ENSDART00000136445
|
si:dkey-16p19.5
|
si:dkey-16p19.5 |
| chr2_-_43168292 | 0.72 |
ENSDART00000132588
|
crema
|
cAMP responsive element modulator a |
| chr12_-_31484677 | 0.72 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr6_+_8176486 | 0.71 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr25_-_12935424 | 0.70 |
ENSDART00000160217
|
ccl39.7
|
chemokine (C-C motif) ligand 39, duplicate 7 |
| chr4_-_12104421 | 0.69 |
ENSDART00000139561
|
mrps33
|
mitochondrial ribosomal protein S33 |
| chr22_-_29242347 | 0.69 |
ENSDART00000040761
|
pvalb7
|
parvalbumin 7 |
| chr24_+_23742690 | 0.68 |
ENSDART00000130162
|
TCF24
|
transcription factor 24 |
| chr7_-_38612230 | 0.68 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr18_+_16246806 | 0.68 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr7_+_21887787 | 0.68 |
ENSDART00000162252
|
pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr13_+_10232695 | 0.67 |
ENSDART00000080805
|
six2a
|
SIX homeobox 2a |
| chr11_-_13126969 | 0.66 |
ENSDART00000169052
ENSDART00000190120 |
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr17_-_30702411 | 0.66 |
ENSDART00000114358
|
zgc:194392
|
zgc:194392 |
| chr20_+_26538137 | 0.64 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr10_-_29120515 | 0.63 |
ENSDART00000162016
ENSDART00000149140 |
zpld1a
|
zona pellucida-like domain containing 1a |
| chr15_-_23647078 | 0.63 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr2_-_24981356 | 0.62 |
ENSDART00000111212
|
nck1a
|
NCK adaptor protein 1a |
| chr4_+_7391110 | 0.60 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr7_+_21887307 | 0.59 |
ENSDART00000052871
|
pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr22_-_17256573 | 0.59 |
ENSDART00000136119
ENSDART00000062891 |
nphs2
|
nephrosis 2, idiopathic, steroid-resistant (podocin) |
| chr9_+_10014817 | 0.58 |
ENSDART00000132065
|
nxph2a
|
neurexophilin 2a |
| chr6_-_31806345 | 0.58 |
ENSDART00000149369
|
si:dkey-209n16.2
|
si:dkey-209n16.2 |
| chr11_+_39672874 | 0.58 |
ENSDART00000046663
ENSDART00000157659 |
camta1b
|
calmodulin binding transcription activator 1b |
| chr21_-_33478164 | 0.57 |
ENSDART00000191542
|
si:ch73-42p12.2
|
si:ch73-42p12.2 |
| chr9_-_18743012 | 0.57 |
ENSDART00000131626
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr11_-_3343463 | 0.56 |
ENSDART00000066177
|
tuba2
|
tubulin, alpha 2 |
| chr9_+_10014514 | 0.55 |
ENSDART00000185590
|
nxph2a
|
neurexophilin 2a |
| chr10_+_22012389 | 0.54 |
ENSDART00000035188
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr21_+_6556635 | 0.53 |
ENSDART00000139598
|
col5a1
|
procollagen, type V, alpha 1 |
| chr15_+_19646902 | 0.53 |
ENSDART00000179822
|
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr24_-_5889878 | 0.52 |
ENSDART00000077951
|
pcolce2b
|
procollagen C-endopeptidase enhancer 2b |
| chr9_+_2002701 | 0.52 |
ENSDART00000082329
|
evx2
|
even-skipped homeobox 2 |
| chr9_-_22361681 | 0.52 |
ENSDART00000129954
|
crygm3
|
crystallin, gamma M3 |
| chr22_-_15280638 | 0.51 |
ENSDART00000063008
|
mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr6_-_25181012 | 0.51 |
ENSDART00000161585
|
lrrc8db
|
leucine rich repeat containing 8 VRAC subunit Db |
| chr7_-_59054322 | 0.51 |
ENSDART00000165390
|
chmp5b
|
charged multivesicular body protein 5b |
| chr11_-_15850794 | 0.50 |
ENSDART00000185946
|
rap1ab
|
RAP1A, member of RAS oncogene family b |
| chr22_+_11535131 | 0.49 |
ENSDART00000113930
|
npb
|
neuropeptide B |
| chr6_-_10964083 | 0.49 |
ENSDART00000181583
|
notum2
|
notum pectinacetylesterase 2 |
| chr23_+_20110086 | 0.48 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr7_+_36552725 | 0.48 |
ENSDART00000173682
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr21_-_12119711 | 0.48 |
ENSDART00000131538
|
celf4
|
CUGBP, Elav-like family member 4 |
| chr4_-_27398385 | 0.47 |
ENSDART00000142117
ENSDART00000150553 ENSDART00000182746 |
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr13_-_11699530 | 0.47 |
ENSDART00000192161
|
slc39a8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr3_+_32112004 | 0.47 |
ENSDART00000105272
|
zgc:173593
|
zgc:173593 |
| chr4_-_8902406 | 0.47 |
ENSDART00000192962
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr25_+_20694177 | 0.47 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr1_+_37752171 | 0.47 |
ENSDART00000183247
ENSDART00000189756 ENSDART00000139448 |
GALNTL6
|
si:ch211-15e22.3 |
| chr20_+_2642855 | 0.46 |
ENSDART00000058775
|
zgc:101562
|
zgc:101562 |
| chr14_+_52563794 | 0.45 |
ENSDART00000168874
|
rpl26
|
ribosomal protein L26 |
| chr7_-_32598383 | 0.45 |
ENSDART00000111055
|
kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr4_+_74943111 | 0.44 |
ENSDART00000004739
|
nup50
|
nucleoporin 50 |
| chr25_-_4148719 | 0.43 |
ENSDART00000112880
ENSDART00000023278 |
fads2
|
fatty acid desaturase 2 |
| chr2_-_55797318 | 0.43 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr23_+_19213472 | 0.42 |
ENSDART00000185985
|
apobec2b
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2b |
| chr10_+_21899753 | 0.42 |
ENSDART00000080155
|
hrh2b
|
histamine receptor H2b |
| chr19_-_42557416 | 0.42 |
ENSDART00000163217
ENSDART00000128278 ENSDART00000162304 ENSDART00000166556 |
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr9_-_23922778 | 0.42 |
ENSDART00000135769
|
col6a3
|
collagen, type VI, alpha 3 |
| chr18_-_21271373 | 0.41 |
ENSDART00000060001
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr22_+_33362552 | 0.41 |
ENSDART00000101580
|
nicn1
|
nicolin 1 |
| chr9_+_29520696 | 0.40 |
ENSDART00000144430
|
fdx1
|
ferredoxin 1 |
| chr18_-_6633984 | 0.40 |
ENSDART00000185241
|
tnni1c
|
troponin I, skeletal, slow c |
| chr17_-_24703778 | 0.40 |
ENSDART00000156061
|
si:ch211-15d5.12
|
si:ch211-15d5.12 |
| chr19_+_17259912 | 0.40 |
ENSDART00000078951
|
MANEAL
|
si:ch211-30b16.2 |
| chr16_+_36126310 | 0.39 |
ENSDART00000166040
ENSDART00000189802 |
sh3bp5b
|
SH3-domain binding protein 5b (BTK-associated) |
| chr10_+_24631976 | 0.38 |
ENSDART00000181928
|
slc46a3
|
solute carrier family 46, member 3 |
| chr1_-_36771712 | 0.38 |
ENSDART00000148386
|
prmt9
|
protein arginine methyltransferase 9 |
| chr23_+_44745317 | 0.38 |
ENSDART00000165654
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr2_+_15776649 | 0.38 |
ENSDART00000156535
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr15_-_8517376 | 0.35 |
ENSDART00000186289
|
npas1
|
neuronal PAS domain protein 1 |
| chr18_+_33237127 | 0.35 |
ENSDART00000133386
|
v2rc2
|
vomeronasal 2 receptor, c2 |
| chr22_-_17611742 | 0.35 |
ENSDART00000144031
|
gpx4a
|
glutathione peroxidase 4a |
| chr1_-_45553602 | 0.34 |
ENSDART00000143664
|
grin2bb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B, genome duplicate b |
| chr1_-_21723329 | 0.34 |
ENSDART00000137138
|
si:ch211-134c9.2
|
si:ch211-134c9.2 |
| chr15_+_17345609 | 0.34 |
ENSDART00000111753
|
vmp1
|
vacuole membrane protein 1 |
| chr23_+_9220436 | 0.34 |
ENSDART00000033663
ENSDART00000139870 |
rps21
|
ribosomal protein S21 |
| chr8_-_28349859 | 0.34 |
ENSDART00000062671
|
tuba8l
|
tubulin, alpha 8 like |
| chr11_+_11271959 | 0.34 |
ENSDART00000190678
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr14_+_33722950 | 0.34 |
ENSDART00000075312
|
apln
|
apelin |
| chr8_+_8893535 | 0.33 |
ENSDART00000139150
|
otud5a
|
OTU deubiquitinase 5a |
| chr11_-_23322182 | 0.33 |
ENSDART00000111289
|
kiss1
|
KiSS-1 metastasis-suppressor |
| chr6_-_39765546 | 0.33 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr7_-_59210882 | 0.33 |
ENSDART00000170330
ENSDART00000158996 |
nagk
|
N-acetylglucosamine kinase |
| chr7_+_30178253 | 0.33 |
ENSDART00000002075
|
si:ch211-107o10.3
|
si:ch211-107o10.3 |
| chr1_+_58091659 | 0.32 |
ENSDART00000144190
|
si:ch211-114l13.1
|
si:ch211-114l13.1 |
| chr21_+_21326038 | 0.32 |
ENSDART00000089898
|
qpctlb
|
glutaminyl-peptide cyclotransferase-like b |
| chr9_-_18742704 | 0.32 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr13_-_22961605 | 0.31 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr11_-_41130239 | 0.30 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr9_+_29585943 | 0.30 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr5_+_51026563 | 0.30 |
ENSDART00000050988
|
gcnt4a
|
glucosaminyl (N-acetyl) transferase 4, core 2, a |
| chr7_+_35238234 | 0.30 |
ENSDART00000178732
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr1_+_33558555 | 0.30 |
ENSDART00000018472
|
chmp2bb
|
charged multivesicular body protein 2Bb |
| chr11_+_35364445 | 0.30 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr2_-_1188707 | 0.29 |
ENSDART00000066378
|
CABZ01084564.1
|
|
| chr8_-_25327809 | 0.29 |
ENSDART00000137242
|
eps8l3b
|
EPS8-like 3b |
| chr2_-_10188598 | 0.29 |
ENSDART00000189122
|
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr25_-_16782394 | 0.29 |
ENSDART00000019413
|
ndufa9a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9a |
| chr4_-_685412 | 0.29 |
ENSDART00000168167
|
rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr5_-_33298352 | 0.28 |
ENSDART00000015076
|
zgc:103692
|
zgc:103692 |
| chr14_-_30141162 | 0.28 |
ENSDART00000053916
|
mtnr1ab
|
melatonin receptor 1A b |
| chr18_-_6634424 | 0.28 |
ENSDART00000062423
ENSDART00000179955 |
tnni1c
|
troponin I, skeletal, slow c |
| chr2_-_20923864 | 0.28 |
ENSDART00000006870
|
ptgs2a
|
prostaglandin-endoperoxide synthase 2a |
| chr17_+_33495194 | 0.28 |
ENSDART00000033691
|
pth2
|
parathyroid hormone 2 |
| chr24_+_25069609 | 0.27 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr9_-_14055959 | 0.27 |
ENSDART00000146675
|
fer1l6
|
fer-1-like family member 6 |
| chr21_-_40799404 | 0.27 |
ENSDART00000147405
ENSDART00000157512 |
zgc:162239
|
zgc:162239 |
| chr11_-_37509001 | 0.27 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr13_+_30172645 | 0.27 |
ENSDART00000137114
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr10_+_28308376 | 0.27 |
ENSDART00000191216
|
ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr1_-_33351780 | 0.26 |
ENSDART00000178996
|
gyg2
|
glycogenin 2 |
| chr23_+_33991509 | 0.26 |
ENSDART00000145161
|
fam50a
|
family with sequence similarity 50, member A |
| chr12_-_6551681 | 0.25 |
ENSDART00000145413
|
si:ch211-253p2.2
|
si:ch211-253p2.2 |
| chr10_+_9159279 | 0.25 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr9_+_31075896 | 0.25 |
ENSDART00000188042
|
clybl
|
citrate lyase beta like |
| chr25_+_25438322 | 0.25 |
ENSDART00000150364
|
LRRC56
|
si:ch211-103e16.5 |
| chr9_-_23990416 | 0.24 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr1_-_26045007 | 0.24 |
ENSDART00000129259
|
gpank1
|
G patch domain and ankyrin repeats 1 |
| chr3_-_18575868 | 0.24 |
ENSDART00000122968
|
aqp8b
|
aquaporin 8b |
| chr16_-_14353567 | 0.24 |
ENSDART00000139859
|
itga10
|
integrin, alpha 10 |
| chr6_-_25180438 | 0.24 |
ENSDART00000159696
|
lrrc8db
|
leucine rich repeat containing 8 VRAC subunit Db |
| chr12_+_638435 | 0.23 |
ENSDART00000152508
|
si:ch211-176g6.1
|
si:ch211-176g6.1 |
| chr10_-_15672862 | 0.23 |
ENSDART00000109231
|
mamdc2b
|
MAM domain containing 2b |
| chr6_+_46259950 | 0.23 |
ENSDART00000032326
|
zgc:162324
|
zgc:162324 |
| chr6_-_15101477 | 0.22 |
ENSDART00000187713
ENSDART00000124132 |
fhl2b
|
four and a half LIM domains 2b |
| chr15_+_5028608 | 0.22 |
ENSDART00000092809
|
abcg1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr13_+_25421531 | 0.22 |
ENSDART00000158093
|
calhm2
|
calcium homeostasis modulator 2 |
| chr21_+_41743493 | 0.21 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr15_+_33991928 | 0.21 |
ENSDART00000170177
|
vwde
|
von Willebrand factor D and EGF domains |
| chr4_-_32456788 | 0.21 |
ENSDART00000151862
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr7_-_44604540 | 0.21 |
ENSDART00000149186
|
tk2
|
thymidine kinase 2, mitochondrial |
| chr24_-_36337395 | 0.21 |
ENSDART00000154402
|
si:ch211-40k21.9
|
si:ch211-40k21.9 |
| chr6_+_21062416 | 0.21 |
ENSDART00000141643
|
si:dkey-91f15.1
|
si:dkey-91f15.1 |
| chr1_-_8020589 | 0.20 |
ENSDART00000143881
|
si:dkeyp-9d4.2
|
si:dkeyp-9d4.2 |
| chr1_-_28089557 | 0.20 |
ENSDART00000161024
ENSDART00000167875 |
snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr1_-_9644630 | 0.20 |
ENSDART00000123725
ENSDART00000161164 |
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr7_-_24364536 | 0.20 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr22_-_5958066 | 0.20 |
ENSDART00000145821
|
si:rp71-36a1.3
|
si:rp71-36a1.3 |
| chr18_-_25276262 | 0.20 |
ENSDART00000131855
|
plin1
|
perilipin 1 |
| chr3_-_36210344 | 0.20 |
ENSDART00000025326
|
csnk1da
|
casein kinase 1, delta a |
| chr22_+_19111444 | 0.20 |
ENSDART00000109655
|
fgf22
|
fibroblast growth factor 22 |
| chr7_+_10351038 | 0.19 |
ENSDART00000173256
|
si:cabz01029535.1
|
si:cabz01029535.1 |
| chr3_+_39579393 | 0.19 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr23_+_30835589 | 0.19 |
ENSDART00000154145
|
npbwr2a
|
neuropeptides B/W receptor 2a |
| chr14_+_22591624 | 0.19 |
ENSDART00000108987
|
gfra4b
|
GDNF family receptor alpha 4b |
| chr1_+_34181581 | 0.19 |
ENSDART00000146042
|
epha6
|
eph receptor A6 |
| chr1_+_58370526 | 0.18 |
ENSDART00000067775
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr14_-_15154695 | 0.18 |
ENSDART00000160677
|
uvssa
|
UV-stimulated scaffold protein A |
Network of associatons between targets according to the STRING database.
First level regulatory network of mecom
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0042941 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.4 | 1.4 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.3 | 0.8 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.3 | 3.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.2 | 1.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 1.2 | GO:0035176 | mammillary body development(GO:0021767) social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.2 | 4.0 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 0.5 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 0.4 | GO:0043388 | cytidine to uridine editing(GO:0016554) positive regulation of DNA binding(GO:0043388) |
| 0.1 | 0.7 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 1.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.5 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.5 | GO:0051591 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.3 | GO:1901006 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.1 | 0.8 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.3 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 1.0 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 1.6 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 0.3 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 2.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.2 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 1.0 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.4 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.9 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.6 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.5 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 1.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.2 | GO:0031652 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.8 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 1.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 2.9 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.3 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0060907 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.2 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.1 | GO:0071312 | response to cocaine(GO:0042220) cellular response to alkaloid(GO:0071312) cellular response to cocaine(GO:0071314) |
| 0.0 | 0.2 | GO:0030194 | acute-phase response(GO:0006953) positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:1903332 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.0 | 0.8 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.7 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.4 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 1.0 | GO:0035138 | pectoral fin morphogenesis(GO:0035138) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.3 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.0 | 0.3 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.3 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.6 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.5 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.8 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.3 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.0 | 1.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.3 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.4 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.7 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.1 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.1 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 4.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.2 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.0 | 1.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 1.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 3.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0072380 | TRC complex(GO:0072380) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0035060 | brahma complex(GO:0035060) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.4 | 1.4 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.4 | 1.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.3 | 0.8 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.2 | 4.0 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 1.5 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 4.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.9 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.3 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.1 | 1.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.6 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.2 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.1 | 0.3 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.3 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 1.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.6 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.4 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.4 | GO:0030955 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.0 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0016496 | substance P receptor activity(GO:0016496) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 3.6 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.7 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) structural constituent of synapse(GO:0098918) |
| 0.0 | 0.0 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |