Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
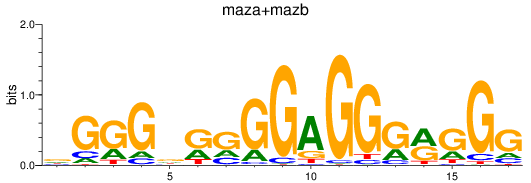
Results for maza+mazb
Z-value: 4.71
Transcription factors associated with maza+mazb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mazb
|
ENSDARG00000063555 | si_ch211-166g5.4 |
|
maza
|
ENSDARG00000087330 | MYC-associated zinc finger protein a (purine-binding transcription factor) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| maza | dr11_v1_chr3_-_21106093_21106093 | -0.43 | 6.7e-02 | Click! |
| si:ch211-166g5.4 | dr11_v1_chr12_-_3773869_3773869 | -0.33 | 1.7e-01 | Click! |
Activity profile of maza+mazb motif
Sorted Z-values of maza+mazb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_6954984 | 23.23 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr3_-_30861177 | 19.24 |
ENSDART00000154811
|
shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr2_-_43168292 | 18.31 |
ENSDART00000132588
|
crema
|
cAMP responsive element modulator a |
| chr18_-_16123222 | 17.87 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr19_-_5125663 | 17.63 |
ENSDART00000005547
ENSDART00000132642 |
gnb3b
|
guanine nucleotide binding protein (G protein), beta polypeptide 3b |
| chr11_+_36989696 | 17.43 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr14_-_30642819 | 16.50 |
ENSDART00000078154
|
npas4a
|
neuronal PAS domain protein 4a |
| chr5_-_67878064 | 15.91 |
ENSDART00000111203
|
tagln3a
|
transgelin 3a |
| chr21_+_1143141 | 15.58 |
ENSDART00000178294
|
CABZ01086139.1
|
|
| chr11_-_44030962 | 14.14 |
ENSDART00000171910
|
FP016005.1
|
|
| chr11_-_11518469 | 13.89 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr6_+_54888493 | 13.70 |
ENSDART00000113331
|
nav1b
|
neuron navigator 1b |
| chr4_-_77557279 | 12.99 |
ENSDART00000180113
|
AL935186.10
|
|
| chr19_+_43123331 | 12.46 |
ENSDART00000187836
|
CABZ01101996.1
|
|
| chr11_-_1291012 | 11.88 |
ENSDART00000158390
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr8_+_22931427 | 11.68 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr17_-_46457622 | 11.50 |
ENSDART00000130215
|
TMEM179 (1 of many)
|
transmembrane protein 179 |
| chr19_-_26863626 | 11.32 |
ENSDART00000145568
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr24_+_37080771 | 11.18 |
ENSDART00000159942
|
kcnc3b
|
potassium voltage-gated channel, Shaw-related subfamily, member 3b |
| chr10_-_44026369 | 10.91 |
ENSDART00000185456
|
crybb1
|
crystallin, beta B1 |
| chr11_-_32723851 | 10.81 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr16_-_17207754 | 10.21 |
ENSDART00000063804
|
wu:fj39g12
|
wu:fj39g12 |
| chr19_-_25301711 | 10.16 |
ENSDART00000175739
|
rims3
|
regulating synaptic membrane exocytosis 3 |
| chr4_+_20255160 | 10.08 |
ENSDART00000188658
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr3_+_37827373 | 10.00 |
ENSDART00000039517
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr18_-_51015718 | 9.94 |
ENSDART00000190698
|
LO018598.1
|
|
| chr5_+_9348284 | 9.72 |
ENSDART00000149417
|
tal2
|
T-cell acute lymphocytic leukemia 2 |
| chr7_-_10606 | 9.70 |
ENSDART00000192650
ENSDART00000186761 |
FO704772.2
|
|
| chr3_-_30625219 | 9.50 |
ENSDART00000151698
|
syt3
|
synaptotagmin III |
| chr12_+_5530247 | 9.35 |
ENSDART00000114637
|
ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr2_-_44183451 | 9.28 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr7_-_72208248 | 9.19 |
ENSDART00000108916
|
zmp:0000001168
|
zmp:0000001168 |
| chr5_-_37935607 | 9.09 |
ENSDART00000141233
ENSDART00000084868 ENSDART00000125857 |
scn4bb
|
sodium channel, voltage-gated, type IV, beta b |
| chr9_+_13714379 | 9.07 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr21_+_5169154 | 9.01 |
ENSDART00000102559
|
zgc:122979
|
zgc:122979 |
| chr13_+_28701233 | 9.00 |
ENSDART00000135931
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr25_-_30047477 | 8.96 |
ENSDART00000164859
|
CR759810.1
|
|
| chr14_+_22172047 | 8.95 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr18_-_46763170 | 8.95 |
ENSDART00000171880
|
dner
|
delta/notch-like EGF repeat containing |
| chr21_+_25660613 | 8.93 |
ENSDART00000134017
|
si:dkey-17e16.15
|
si:dkey-17e16.15 |
| chr19_-_1961024 | 8.86 |
ENSDART00000108784
|
mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr13_+_1100197 | 8.83 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr19_-_5254699 | 8.78 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr23_-_35756115 | 8.68 |
ENSDART00000043429
|
jph2
|
junctophilin 2 |
| chr25_+_37366698 | 8.67 |
ENSDART00000165400
ENSDART00000192589 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr2_+_22694382 | 8.58 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr1_+_37391141 | 8.53 |
ENSDART00000083593
ENSDART00000168647 |
sparcl1
|
SPARC-like 1 |
| chr4_-_17793152 | 8.53 |
ENSDART00000134080
|
mybpc1
|
myosin binding protein C, slow type |
| chr3_+_22327738 | 8.52 |
ENSDART00000055675
|
gh1
|
growth hormone 1 |
| chr3_+_32425202 | 8.47 |
ENSDART00000156464
|
prr12b
|
proline rich 12b |
| chr2_-_5014939 | 8.44 |
ENSDART00000164506
|
dlg1l
|
discs, large (Drosophila) homolog 1, like |
| chr3_+_54168007 | 8.33 |
ENSDART00000109894
|
olfm2a
|
olfactomedin 2a |
| chr3_-_28120092 | 8.31 |
ENSDART00000151143
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr4_+_5537101 | 8.30 |
ENSDART00000008692
|
si:dkey-14d8.7
|
si:dkey-14d8.7 |
| chr4_-_6459863 | 8.24 |
ENSDART00000138367
|
foxp2
|
forkhead box P2 |
| chr6_+_38381957 | 8.22 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr4_+_2619132 | 8.14 |
ENSDART00000128807
|
gpr22a
|
G protein-coupled receptor 22a |
| chr24_-_38110779 | 8.07 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr20_-_25518488 | 8.01 |
ENSDART00000186993
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr7_-_58098814 | 7.97 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr13_+_27073901 | 7.94 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr21_-_37889727 | 7.90 |
ENSDART00000163612
ENSDART00000180958 |
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr6_+_54358529 | 7.89 |
ENSDART00000153704
|
anks1ab
|
ankyrin repeat and sterile alpha motif domain containing 1Ab |
| chr2_-_44183613 | 7.89 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr22_-_21845685 | 7.81 |
ENSDART00000105564
|
aes
|
amino-terminal enhancer of split |
| chr1_-_31505144 | 7.81 |
ENSDART00000087115
|
rims1b
|
regulating synaptic membrane exocytosis 1b |
| chr24_+_36452927 | 7.65 |
ENSDART00000168181
|
zgc:114120
|
zgc:114120 |
| chr22_+_32120156 | 7.64 |
ENSDART00000149666
|
dock3
|
dedicator of cytokinesis 3 |
| chr10_+_22724059 | 7.63 |
ENSDART00000136123
|
kdm6bb
|
lysine (K)-specific demethylase 6B, b |
| chr12_-_22039350 | 7.58 |
ENSDART00000153187
|
thrab
|
thyroid hormone receptor alpha b |
| chr10_-_37337579 | 7.52 |
ENSDART00000147705
|
omga
|
oligodendrocyte myelin glycoprotein a |
| chr1_+_12009673 | 7.48 |
ENSDART00000080100
|
slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr3_-_36115339 | 7.45 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr11_-_44543082 | 7.45 |
ENSDART00000099568
|
gpr137bb
|
G protein-coupled receptor 137Bb |
| chr12_-_45349849 | 7.38 |
ENSDART00000183036
|
CABZ01068367.1
|
Danio rerio uncharacterized LOC100332446 (LOC100332446), mRNA. |
| chr4_+_55758103 | 7.34 |
ENSDART00000185964
|
CT583728.23
|
|
| chr9_-_49531762 | 7.33 |
ENSDART00000121875
|
xirp2b
|
xin actin binding repeat containing 2b |
| chr7_+_427503 | 7.27 |
ENSDART00000185942
|
NRXN2 (1 of many)
|
neurexin 2 |
| chr22_+_38159823 | 7.21 |
ENSDART00000104527
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr13_-_21739142 | 7.19 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr14_-_7207961 | 7.18 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr20_+_15982482 | 7.16 |
ENSDART00000020999
|
angptl1a
|
angiopoietin-like 1a |
| chr23_+_14217508 | 7.13 |
ENSDART00000143618
|
birc7
|
baculoviral IAP repeat containing 7 |
| chr10_-_35542071 | 7.10 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr7_+_528593 | 7.08 |
ENSDART00000091955
|
nrxn2b
|
neurexin 2b |
| chr15_-_12500938 | 7.04 |
ENSDART00000159627
|
scn4ba
|
sodium channel, voltage-gated, type IV, beta a |
| chr24_-_1985007 | 7.02 |
ENSDART00000189870
|
PARD3 (1 of many)
|
par-3 family cell polarity regulator |
| chr2_-_9646857 | 7.01 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr3_-_3209432 | 6.97 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr14_-_2369849 | 6.90 |
ENSDART00000180422
ENSDART00000189731 ENSDART00000111748 |
pcdhb
|
protocadherin b |
| chr15_-_12113045 | 6.85 |
ENSDART00000159879
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr8_+_46418996 | 6.81 |
ENSDART00000144285
|
si:ch211-196g2.4
|
si:ch211-196g2.4 |
| chr8_-_14052349 | 6.80 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr8_-_22660678 | 6.75 |
ENSDART00000181258
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr1_-_22370660 | 6.74 |
ENSDART00000127506
|
si:ch73-380n15.2
|
si:ch73-380n15.2 |
| chr18_+_21122818 | 6.72 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr19_-_6193448 | 6.70 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr25_-_12788370 | 6.70 |
ENSDART00000158551
|
slc7a5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr3_-_52614747 | 6.65 |
ENSDART00000154365
|
trim35-13
|
tripartite motif containing 35-13 |
| chr20_+_13969414 | 6.63 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr25_+_19954576 | 6.61 |
ENSDART00000149335
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr18_+_12058403 | 6.60 |
ENSDART00000140854
ENSDART00000193632 ENSDART00000190519 ENSDART00000190685 ENSDART00000112671 |
bicd1a
|
bicaudal D homolog 1a |
| chr13_-_349952 | 6.56 |
ENSDART00000133731
ENSDART00000140326 ENSDART00000189389 ENSDART00000185865 ENSDART00000109634 ENSDART00000147058 ENSDART00000142695 |
si:ch1073-291c23.2
|
si:ch1073-291c23.2 |
| chr7_-_71837213 | 6.50 |
ENSDART00000168645
ENSDART00000160512 |
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr16_-_36798783 | 6.47 |
ENSDART00000145697
|
calb1
|
calbindin 1 |
| chr23_+_6232895 | 6.46 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr19_-_9760994 | 6.37 |
ENSDART00000139727
|
si:dkey-14o18.2
|
si:dkey-14o18.2 |
| chr3_+_37824268 | 6.32 |
ENSDART00000137038
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr2_+_55199721 | 6.29 |
ENSDART00000016143
|
zmp:0000000521
|
zmp:0000000521 |
| chr1_-_45647846 | 6.29 |
ENSDART00000186881
|
BX511120.1
|
|
| chr1_+_44491077 | 6.24 |
ENSDART00000073736
|
rtn4rl2a
|
reticulon 4 receptor-like 2 a |
| chr7_-_6604623 | 6.24 |
ENSDART00000172874
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr19_-_32804535 | 6.21 |
ENSDART00000175613
ENSDART00000052098 |
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr4_+_1146346 | 6.14 |
ENSDART00000166467
|
chrm2a
|
cholinergic receptor, muscarinic 2a |
| chr14_-_1535430 | 6.11 |
ENSDART00000179228
|
CABZ01109481.1
|
|
| chr20_-_14665002 | 6.07 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr15_-_2188332 | 6.00 |
ENSDART00000138941
ENSDART00000009564 |
shox2
|
short stature homeobox 2 |
| chr6_+_22597362 | 5.94 |
ENSDART00000131242
|
cygb2
|
cytoglobin 2 |
| chr11_+_39969048 | 5.94 |
ENSDART00000193693
|
per3
|
period circadian clock 3 |
| chr12_+_48744016 | 5.86 |
ENSDART00000175518
|
LO017648.1
|
|
| chr25_-_19395476 | 5.80 |
ENSDART00000182622
|
map1ab
|
microtubule-associated protein 1Ab |
| chr12_+_32729470 | 5.76 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr21_-_21373242 | 5.72 |
ENSDART00000079629
|
ppm1nb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Nb (putative) |
| chr11_-_97817 | 5.71 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr6_+_59029485 | 5.68 |
ENSDART00000050140
|
CABZ01088367.1
|
|
| chr13_+_38430466 | 5.65 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr4_-_1360495 | 5.65 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr24_-_38579066 | 5.64 |
ENSDART00000099869
|
slc17a7b
|
solute carrier family 17 (vesicular glutamate transporter), member 7b |
| chr16_-_22683038 | 5.62 |
ENSDART00000138130
|
s100t
|
S100 calcium binding protein T |
| chr20_+_50115335 | 5.57 |
ENSDART00000031139
|
slc24a4b
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4b |
| chr7_+_27977065 | 5.50 |
ENSDART00000089574
|
tub
|
tubby bipartite transcription factor |
| chr8_-_51293265 | 5.46 |
ENSDART00000127875
ENSDART00000181145 |
bmp1a
|
bone morphogenetic protein 1a |
| chr19_+_9032073 | 5.43 |
ENSDART00000127755
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr16_-_12914288 | 5.41 |
ENSDART00000184221
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr4_+_20263097 | 5.38 |
ENSDART00000138820
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr25_+_35706493 | 5.32 |
ENSDART00000176741
|
kif21a
|
kinesin family member 21A |
| chr8_-_20230559 | 5.31 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr3_-_10634438 | 5.27 |
ENSDART00000093037
ENSDART00000130761 ENSDART00000156617 |
map2k4a
|
mitogen-activated protein kinase kinase 4a |
| chr15_+_47903864 | 5.16 |
ENSDART00000063835
|
otx5
|
orthodenticle homolog 5 |
| chr24_-_31942656 | 5.15 |
ENSDART00000180308
ENSDART00000016879 |
c1ql3a
|
complement component 1, q subcomponent-like 3a |
| chr24_-_6501211 | 5.14 |
ENSDART00000186241
ENSDART00000109040 ENSDART00000136154 |
gpr158a
|
G protein-coupled receptor 158a |
| chr8_+_13849999 | 5.11 |
ENSDART00000143784
|
doc2d
|
double C2-like domains, delta |
| chr16_-_27640995 | 5.10 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr5_+_5689476 | 5.08 |
ENSDART00000022729
|
unm_sa808
|
un-named sa808 |
| chr7_-_32598383 | 5.03 |
ENSDART00000111055
|
kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr20_+_50184934 | 5.02 |
ENSDART00000158388
|
slc24a4b
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4b |
| chr8_-_5847533 | 5.00 |
ENSDART00000192489
|
CABZ01102147.1
|
|
| chr12_-_13886952 | 5.00 |
ENSDART00000110503
|
adam11
|
ADAM metallopeptidase domain 11 |
| chr21_+_11244068 | 4.98 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr15_-_28618502 | 4.96 |
ENSDART00000086902
|
slc6a4a
|
solute carrier family 6 (neurotransmitter transporter), member 4a |
| chr15_-_5467477 | 4.94 |
ENSDART00000123839
|
arrb1
|
arrestin, beta 1 |
| chr6_+_58492201 | 4.93 |
ENSDART00000156375
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr19_+_9533008 | 4.93 |
ENSDART00000104607
|
fam131ba
|
family with sequence similarity 131, member Ba |
| chr20_-_51727860 | 4.86 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr3_-_28075756 | 4.79 |
ENSDART00000122037
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr1_-_29045426 | 4.79 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr7_+_36539124 | 4.78 |
ENSDART00000173653
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr15_-_33925851 | 4.75 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr1_+_45351890 | 4.75 |
ENSDART00000145486
|
si:ch211-243a20.3
|
si:ch211-243a20.3 |
| chr2_-_50372467 | 4.73 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein like 2b |
| chr1_+_6307305 | 4.71 |
ENSDART00000183870
|
LO018629.1
|
|
| chr21_+_26697536 | 4.70 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr18_-_41219790 | 4.70 |
ENSDART00000162387
ENSDART00000193753 |
zbtb38
|
zinc finger and BTB domain containing 38 |
| chr2_+_48073972 | 4.68 |
ENSDART00000186442
|
klf6b
|
Kruppel-like factor 6b |
| chr21_+_41649336 | 4.67 |
ENSDART00000164694
ENSDART00000181539 |
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr25_+_336503 | 4.66 |
ENSDART00000160395
|
CU929262.1
|
|
| chr15_-_37543591 | 4.65 |
ENSDART00000180400
|
kmt2bb
|
lysine (K)-specific methyltransferase 2Bb |
| chr5_-_68022631 | 4.63 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr10_-_9115383 | 4.62 |
ENSDART00000139324
|
si:dkeyp-41f9.3
|
si:dkeyp-41f9.3 |
| chr20_-_34754617 | 4.62 |
ENSDART00000148066
|
znf395b
|
zinc finger protein 395b |
| chr1_+_44127292 | 4.62 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr8_+_14855471 | 4.58 |
ENSDART00000134775
|
si:dkey-21p1.3
|
si:dkey-21p1.3 |
| chr2_-_4787566 | 4.57 |
ENSDART00000160663
ENSDART00000157808 |
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr11_-_21304452 | 4.53 |
ENSDART00000163008
|
RASSF5
|
si:dkey-85p17.3 |
| chr23_-_31693309 | 4.47 |
ENSDART00000146327
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr22_+_883678 | 4.47 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr10_-_7974155 | 4.47 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr2_-_5776030 | 4.46 |
ENSDART00000133851
|
nyap2b
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2b |
| chr20_+_33532296 | 4.46 |
ENSDART00000153153
|
kcnf1a
|
potassium voltage-gated channel, subfamily F, member 1a |
| chr13_-_638485 | 4.41 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr19_-_41213718 | 4.39 |
ENSDART00000077121
|
pdk4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr18_-_46464501 | 4.38 |
ENSDART00000040669
|
sphkap
|
SPHK1 interactor, AKAP domain containing |
| chr16_+_24842662 | 4.36 |
ENSDART00000157333
|
si:dkey-79d12.6
|
si:dkey-79d12.6 |
| chr9_-_704667 | 4.34 |
ENSDART00000147092
|
cflarb
|
CASP8 and FADD-like apoptosis regulator b |
| chr14_+_40685469 | 4.34 |
ENSDART00000172839
|
tnmd
|
tenomodulin |
| chr8_+_3496204 | 4.32 |
ENSDART00000085993
|
pxnb
|
paxillin b |
| chr21_+_45841731 | 4.30 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr8_+_48603398 | 4.30 |
ENSDART00000074900
|
zgc:195023
|
zgc:195023 |
| chr5_-_5831037 | 4.30 |
ENSDART00000112856
|
zmp:0000000846
|
zmp:0000000846 |
| chr5_+_43427841 | 4.30 |
ENSDART00000177895
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr6_-_4228640 | 4.28 |
ENSDART00000162497
ENSDART00000179923 |
trak2
|
trafficking protein, kinesin binding 2 |
| chr2_-_38125657 | 4.27 |
ENSDART00000143433
|
cbln12
|
cerebellin 12 |
| chr8_+_16990120 | 4.24 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr12_+_32388860 | 4.23 |
ENSDART00000075568
|
nog3
|
noggin 3 |
| chr2_+_7192966 | 4.20 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr14_+_29550323 | 4.20 |
ENSDART00000157943
|
TENM2
|
si:dkey-34l15.2 |
| chr2_+_33052360 | 4.17 |
ENSDART00000134651
|
rnf220a
|
ring finger protein 220a |
| chr24_-_29729799 | 4.17 |
ENSDART00000181250
|
si:ch73-383g2.1
|
si:ch73-383g2.1 |
| chr5_-_24642026 | 4.17 |
ENSDART00000139522
|
si:ch211-106a19.1
|
si:ch211-106a19.1 |
| chr9_-_3834686 | 4.15 |
ENSDART00000141492
|
myo3b
|
myosin IIIB |
Network of associatons between targets according to the STRING database.
First level regulatory network of maza+mazb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 17.4 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 3.3 | 16.5 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 3.2 | 13.0 | GO:0006837 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 3.0 | 8.9 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 2.5 | 10.1 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 2.5 | 7.5 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 2.5 | 7.5 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 2.5 | 7.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 1.8 | 8.9 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 1.8 | 8.8 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 1.7 | 8.5 | GO:0050996 | positive regulation of lipid catabolic process(GO:0050996) |
| 1.6 | 6.5 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 1.6 | 8.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 1.6 | 34.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 1.3 | 3.8 | GO:0042368 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 1.2 | 4.9 | GO:0060300 | microglial cell activation(GO:0001774) regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 1.1 | 5.7 | GO:0016322 | neuron remodeling(GO:0016322) |
| 1.1 | 6.6 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 1.0 | 4.1 | GO:0010226 | response to lithium ion(GO:0010226) |
| 1.0 | 4.0 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.9 | 4.7 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.9 | 5.5 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.9 | 5.5 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.9 | 6.2 | GO:0036268 | swimming(GO:0036268) |
| 0.9 | 3.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.9 | 4.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.9 | 2.6 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.8 | 7.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.8 | 7.6 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.8 | 3.8 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.7 | 3.7 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.7 | 4.4 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.7 | 3.6 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.7 | 4.3 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.7 | 6.4 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.7 | 5.6 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.6 | 6.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.6 | 2.5 | GO:0090244 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.6 | 1.9 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.6 | 3.6 | GO:0015846 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.6 | 1.8 | GO:0001783 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) negative regulation of B cell proliferation(GO:0030889) negative regulation of B cell activation(GO:0050869) lymphocyte apoptotic process(GO:0070227) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.6 | 1.8 | GO:0035992 | tendon formation(GO:0035992) |
| 0.6 | 3.5 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.6 | 3.4 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.6 | 4.0 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.6 | 2.8 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.5 | 5.5 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.5 | 3.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.5 | 8.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.5 | 16.8 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.5 | 2.5 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.5 | 5.3 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.5 | 6.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.5 | 13.8 | GO:0099633 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.5 | 4.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.4 | 1.8 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.4 | 4.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.4 | 10.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.4 | 3.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.4 | 9.6 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.4 | 7.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.4 | 2.8 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.4 | 1.2 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.4 | 5.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.4 | 4.2 | GO:0035844 | cloaca development(GO:0035844) |
| 0.4 | 3.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.4 | 6.2 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.4 | 10.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.4 | 7.9 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.4 | 16.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.3 | 5.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 3.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.3 | 8.4 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.3 | 6.7 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.3 | 3.7 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.3 | 2.9 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.3 | 9.1 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.3 | 1.6 | GO:0031650 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.3 | 8.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.3 | 4.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.3 | 10.8 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.3 | 9.7 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.3 | 5.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 2.8 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.3 | 3.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.3 | 2.8 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.3 | 1.9 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.3 | 2.6 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.3 | 5.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.3 | 3.9 | GO:0031060 | regulation of histone methylation(GO:0031060) |
| 0.3 | 9.5 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.3 | 6.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 15.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.2 | 30.6 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.2 | 10.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 13.9 | GO:0001764 | neuron migration(GO:0001764) |
| 0.2 | 5.1 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.2 | 4.8 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.2 | 2.6 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.2 | 4.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.2 | 0.2 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.2 | 3.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 0.9 | GO:0090579 | establishment of mitotic sister chromatid cohesion(GO:0034087) establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 4.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.2 | 3.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 3.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 9.3 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.2 | 2.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 7.1 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.2 | 15.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.2 | 4.0 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 16.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 0.6 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.2 | 0.4 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.2 | 35.8 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.2 | 0.8 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.2 | 3.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.7 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.2 | 1.5 | GO:0070293 | renal absorption(GO:0070293) |
| 0.2 | 1.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 5.2 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.2 | 8.2 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.2 | 2.0 | GO:0030534 | adult behavior(GO:0030534) |
| 0.2 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 7.4 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 5.7 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.1 | 4.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 3.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 15.3 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 3.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 3.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 3.6 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.1 | 4.3 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 1.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 2.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 1.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 8.5 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.1 | 2.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 1.8 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.7 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 4.4 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 1.2 | GO:0071384 | response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.1 | 12.4 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.1 | 4.8 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 0.9 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 5.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 4.3 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 5.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 4.0 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.1 | 7.0 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.1 | 1.6 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 1.5 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 4.8 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 4.5 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.1 | 3.2 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.1 | 1.8 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 15.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 1.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 2.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.7 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 1.7 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 3.1 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 10.4 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.1 | 2.5 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 0.1 | 1.2 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 0.7 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 0.8 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 3.2 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.1 | 1.8 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 3.5 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 3.8 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 1.5 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 2.2 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 3.5 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 2.1 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.8 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 8.7 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.5 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 3.3 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.2 | GO:0007183 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 1.9 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 3.4 | GO:0014032 | neural crest cell development(GO:0014032) |
| 0.0 | 5.0 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 6.8 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 1.1 | GO:0048268 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) clathrin coat assembly(GO:0048268) |
| 0.0 | 1.0 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.5 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 4.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.5 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 1.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.3 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.0 | 0.1 | GO:0010002 | cardioblast differentiation(GO:0010002) negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 2.9 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 1.5 | GO:0007018 | microtubule-based movement(GO:0007018) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 18.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 1.7 | 8.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.3 | 6.5 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 1.0 | 4.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 1.0 | 28.0 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.9 | 5.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.8 | 5.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.8 | 17.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.7 | 5.4 | GO:0016586 | RSC complex(GO:0016586) |
| 0.7 | 6.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.6 | 11.2 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.6 | 32.9 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.6 | 4.7 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.6 | 8.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.6 | 6.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.6 | 18.9 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.5 | 7.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.5 | 8.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.4 | 3.5 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.4 | 6.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.4 | 7.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.4 | 5.6 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 2.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.4 | 20.5 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.3 | 19.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 21.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 3.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 4.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 1.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.3 | 3.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 9.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.2 | 3.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 2.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 3.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 2.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 4.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 2.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 4.0 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 7.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 16.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 10.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 1.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 5.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 5.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 2.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.2 | 15.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 3.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 4.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 12.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 4.3 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 3.1 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 1.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 7.7 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 1.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 6.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 3.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 4.7 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 4.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 2.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 9.4 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 2.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 4.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 3.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 4.5 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 2.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 4.3 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.0 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 9.8 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 1.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 3.4 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 14.1 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 40.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 3.9 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 17.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 4.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 88.6 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 20.3 | GO:0005829 | cytosol(GO:0005829) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 17.4 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 2.9 | 26.0 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 2.8 | 8.5 | GO:0070186 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 2.2 | 6.7 | GO:0042166 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 2.2 | 6.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.9 | 7.5 | GO:0071253 | connexin binding(GO:0071253) |
| 1.7 | 5.0 | GO:0005335 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 1.4 | 16.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 1.3 | 3.8 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 1.2 | 19.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.2 | 6.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 1.2 | 5.9 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 1.1 | 5.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 1.0 | 15.1 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.9 | 6.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.8 | 5.9 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.8 | 6.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.8 | 7.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.8 | 35.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.7 | 5.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.7 | 6.4 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.7 | 20.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.7 | 3.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.6 | 4.3 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.6 | 2.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.6 | 3.6 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.6 | 5.6 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.6 | 31.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.6 | 6.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.6 | 18.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.5 | 4.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.5 | 7.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.5 | 2.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.5 | 4.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.5 | 8.8 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.4 | 8.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.4 | 8.0 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.4 | 5.0 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.4 | 2.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.4 | 16.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.3 | 10.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 16.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.3 | 7.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.3 | 13.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 16.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.3 | 3.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 2.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 7.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.3 | 6.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 1.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 3.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.3 | 1.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 10.0 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.3 | 5.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 10.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.3 | 2.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 1.0 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.3 | 2.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 2.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 3.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 7.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 0.7 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.2 | 9.4 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.2 | 4.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 1.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 4.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 8.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 2.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 11.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 4.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 5.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 3.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.2 | 4.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 5.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 4.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 3.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 3.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.2 | 3.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.2 | 1.5 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.2 | 17.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.2 | 8.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 5.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.2 | 10.2 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.2 | 1.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 0.9 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 1.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 5.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 1.5 | GO:0042562 | hormone binding(GO:0042562) |
| 0.1 | 1.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 10.3 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.1 | 1.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 3.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 10.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 7.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 15.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 4.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 1.1 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 3.1 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 3.4 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 7.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 3.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 5.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 1.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 15.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 5.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 3.1 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 1.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.3 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 3.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 2.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 2.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 5.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 4.5 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.1 | 5.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 10.5 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 0.9 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 4.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.1 | 4.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 15.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 19.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 9.3 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 0.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 5.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 14.2 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.5 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 2.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 12.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 4.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 7.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.5 | 8.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.3 | 5.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 3.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 8.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.2 | 6.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 6.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.2 | 2.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 5.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 9.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 2.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 3.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 3.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 17.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.9 | 6.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.7 | 8.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.7 | 13.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.7 | 8.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.6 | 8.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.6 | 13.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.6 | 13.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.5 | 3.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.5 | 6.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.5 | 4.0 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.5 | 7.9 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.4 | 4.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.4 | 12.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.4 | 3.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 4.9 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.3 | 8.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 3.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 15.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 2.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 3.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.2 | 2.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 3.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 2.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 1.7 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.2 | 7.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 3.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 1.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 5.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.9 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 3.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 3.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 3.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.9 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |