Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
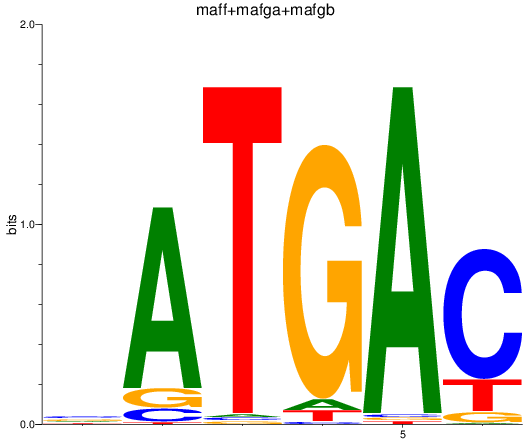
Results for maff+mafga+mafgb
Z-value: 1.36
Transcription factors associated with maff+mafga+mafgb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafga
|
ENSDARG00000018109 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ga |
|
maff
|
ENSDARG00000028957 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
|
mafgb
|
ENSDARG00000100097 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
|
maff
|
ENSDARG00000111050 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mafga | dr11_v1_chr3_-_55404985_55404985 | 0.92 | 2.2e-08 | Click! |
| mafgb | dr11_v1_chr11_+_45323455_45323455 | 0.91 | 8.9e-08 | Click! |
| maff | dr11_v1_chr12_-_19346678_19346678 | 0.56 | 1.3e-02 | Click! |
Activity profile of maff+mafga+mafgb motif
Sorted Z-values of maff+mafga+mafgb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_32526799 | 14.46 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr15_-_34418525 | 5.84 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr22_+_19289970 | 4.84 |
ENSDART00000137976
ENSDART00000132386 |
si:dkey-21e2.15
|
si:dkey-21e2.15 |
| chr6_-_35446110 | 4.52 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr20_+_35473288 | 3.87 |
ENSDART00000047195
|
mep1a.1
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 1 |
| chr8_+_554531 | 3.84 |
ENSDART00000193623
|
FO704758.2
|
|
| chr19_-_703898 | 3.73 |
ENSDART00000181096
ENSDART00000121462 |
slc6a19a.2
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 2 |
| chr12_-_32421046 | 3.70 |
ENSDART00000075567
|
enpp7.1
|
ectonucleotide pyrophosphatase/phosphodiesterase 7, tandem duplicate 1 |
| chr15_-_44512461 | 3.19 |
ENSDART00000155456
|
gria4a
|
glutamate receptor, ionotropic, AMPA 4a |
| chr3_-_39171968 | 3.16 |
ENSDART00000154494
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr21_-_45613564 | 3.10 |
ENSDART00000160324
|
LO018363.1
|
|
| chr21_-_43606502 | 3.09 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr9_+_1162216 | 3.08 |
ENSDART00000165295
|
stk24a
|
serine/threonine kinase 24a (STE20 homolog, yeast) |
| chr8_+_53452681 | 3.04 |
ENSDART00000166705
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr22_+_19290199 | 3.00 |
ENSDART00000148173
|
si:dkey-21e2.15
|
si:dkey-21e2.15 |
| chr15_-_34408777 | 2.85 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr10_+_29698467 | 2.81 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr10_-_25699454 | 2.73 |
ENSDART00000064376
|
sod1
|
superoxide dismutase 1, soluble |
| chr2_-_32505091 | 2.71 |
ENSDART00000141884
ENSDART00000056639 |
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr9_-_43071519 | 2.68 |
ENSDART00000109099
|
ttn.2
|
titin, tandem duplicate 2 |
| chr12_-_31422433 | 2.66 |
ENSDART00000186075
ENSDART00000153172 ENSDART00000066256 |
vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr5_-_7897316 | 2.65 |
ENSDART00000160743
ENSDART00000160912 |
opn4xb
|
opsin 4xb |
| chr16_-_12173554 | 2.64 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr3_-_32169754 | 2.63 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr11_-_11458208 | 2.62 |
ENSDART00000005485
|
krt93
|
keratin 93 |
| chr14_+_31721171 | 2.59 |
ENSDART00000173034
|
adgrg4a
|
adhesion G protein-coupled receptor G4a |
| chr16_+_10777116 | 2.53 |
ENSDART00000190902
|
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr9_-_42861080 | 2.53 |
ENSDART00000193688
|
ttn.1
|
titin, tandem duplicate 1 |
| chr8_+_46939391 | 2.50 |
ENSDART00000146631
|
espn
|
espin |
| chr22_+_19277361 | 2.50 |
ENSDART00000190075
ENSDART00000141097 ENSDART00000136943 |
si:dkey-21e2.13
|
si:dkey-21e2.13 |
| chr23_-_26535875 | 2.49 |
ENSDART00000135988
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr9_-_23922011 | 2.42 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr8_+_53423408 | 2.38 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr8_-_46894362 | 2.37 |
ENSDART00000111124
|
acot7
|
acyl-CoA thioesterase 7 |
| chr13_+_28675686 | 2.35 |
ENSDART00000027213
|
inaa
|
internexin neuronal intermediate filament protein, alpha a |
| chr14_-_8890437 | 2.34 |
ENSDART00000167242
|
ATG2A
|
si:ch73-45o6.2 |
| chr7_+_49865049 | 2.32 |
ENSDART00000164165
|
slc1a2a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2a |
| chr23_-_270847 | 2.30 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr25_+_37443194 | 2.27 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr19_+_43563179 | 2.23 |
ENSDART00000151478
|
cd164l2
|
CD164 sialomucin-like 2 |
| chr17_+_27134806 | 2.20 |
ENSDART00000151901
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr21_-_28340977 | 2.15 |
ENSDART00000141629
|
nrxn2a
|
neurexin 2a |
| chr6_+_27667359 | 2.14 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr7_+_36539124 | 2.11 |
ENSDART00000173653
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr14_-_2355833 | 2.11 |
ENSDART00000157677
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.6 |
| chr24_-_17023392 | 2.03 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr2_+_20332044 | 2.02 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr14_+_81919 | 2.01 |
ENSDART00000171444
|
stag3
|
stromal antigen 3 |
| chr7_-_878473 | 1.99 |
ENSDART00000173567
|
si:cabz01080528.1
|
si:cabz01080528.1 |
| chr20_-_27864964 | 1.95 |
ENSDART00000153311
|
syndig1l
|
synapse differentiation inducing 1-like |
| chr10_-_27049170 | 1.93 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr16_-_17175731 | 1.93 |
ENSDART00000183057
|
opn9
|
opsin 9 |
| chr21_+_30194904 | 1.91 |
ENSDART00000137023
ENSDART00000078403 |
BRD8
|
si:ch211-59d17.3 |
| chr7_+_25036188 | 1.91 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr19_-_19339285 | 1.87 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr19_-_1961024 | 1.85 |
ENSDART00000108784
|
mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr7_+_67486807 | 1.85 |
ENSDART00000159989
|
cpne7
|
copine VII |
| chr22_+_2254972 | 1.84 |
ENSDART00000144906
|
znf1157
|
zinc finger protein 1157 |
| chr5_-_26950374 | 1.81 |
ENSDART00000050542
|
htra4
|
HtrA serine peptidase 4 |
| chr8_+_25351863 | 1.80 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr18_+_7286788 | 1.78 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr8_+_50983551 | 1.78 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr23_-_27345425 | 1.78 |
ENSDART00000022042
ENSDART00000191870 |
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr23_+_2760573 | 1.77 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr23_+_13375359 | 1.77 |
ENSDART00000151947
|
ZNF512B
|
si:dkey-256k13.2 |
| chr19_-_29887629 | 1.76 |
ENSDART00000066123
|
kpna6
|
karyopherin alpha 6 (importin alpha 7) |
| chr3_-_41795917 | 1.75 |
ENSDART00000182662
|
grifin
|
galectin-related inter-fiber protein |
| chr10_+_1638876 | 1.74 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr21_+_9666347 | 1.73 |
ENSDART00000163853
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr17_-_22062364 | 1.72 |
ENSDART00000114470
|
ttbk1b
|
tau tubulin kinase 1b |
| chr1_+_34203817 | 1.70 |
ENSDART00000191432
ENSDART00000046094 |
arl6
|
ADP-ribosylation factor-like 6 |
| chr7_-_18730474 | 1.70 |
ENSDART00000170374
|
si:dkey-38l22.2
|
si:dkey-38l22.2 |
| chr6_-_51101834 | 1.68 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr23_-_26536055 | 1.68 |
ENSDART00000182719
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr23_+_216012 | 1.67 |
ENSDART00000181115
ENSDART00000004678 ENSDART00000190439 ENSDART00000189322 |
PDZD4
|
si:ch73-162j3.4 |
| chr7_+_23515966 | 1.67 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr14_+_33413980 | 1.66 |
ENSDART00000052780
ENSDART00000124437 ENSDART00000173327 |
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr7_+_25059845 | 1.63 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr3_+_15907458 | 1.63 |
ENSDART00000163525
|
mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr5_+_27137473 | 1.62 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr2_-_24996441 | 1.62 |
ENSDART00000144795
|
slc35g2a
|
solute carrier family 35, member G2a |
| chr3_-_60142530 | 1.61 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr19_-_10425140 | 1.58 |
ENSDART00000145319
|
si:ch211-171h4.3
|
si:ch211-171h4.3 |
| chr7_-_42206720 | 1.57 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr12_-_3903886 | 1.57 |
ENSDART00000184214
ENSDART00000041082 |
gdpd3b
|
glycerophosphodiester phosphodiesterase domain containing 3b |
| chr21_+_18353703 | 1.56 |
ENSDART00000181396
ENSDART00000166359 |
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr17_-_37195163 | 1.53 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr2_+_55916911 | 1.52 |
ENSDART00000189483
ENSDART00000183647 ENSDART00000083470 |
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr5_-_18513950 | 1.49 |
ENSDART00000145878
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr6_-_10780698 | 1.49 |
ENSDART00000151714
|
gpr155b
|
G protein-coupled receptor 155b |
| chr23_-_4915118 | 1.48 |
ENSDART00000060714
|
atp6ap1a
|
ATPase H+ transporting accessory protein 1a |
| chr24_-_39771667 | 1.48 |
ENSDART00000181867
|
GFOD1
|
si:ch211-276f18.2 |
| chr9_+_27411502 | 1.46 |
ENSDART00000143994
|
si:dkey-193n17.9
|
si:dkey-193n17.9 |
| chr14_-_24110707 | 1.46 |
ENSDART00000133522
ENSDART00000123152 |
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr13_+_36146415 | 1.44 |
ENSDART00000140301
|
TTC9
|
si:ch211-259k16.3 |
| chr12_+_16168342 | 1.44 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr11_-_41130239 | 1.44 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr17_+_20204122 | 1.43 |
ENSDART00000078672
|
gnrh3
|
gonadotropin-releasing hormone 3 |
| chr3_-_57744323 | 1.41 |
ENSDART00000101829
|
lgals3bpb
|
lectin, galactoside-binding, soluble, 3 binding protein b |
| chr10_+_20113830 | 1.40 |
ENSDART00000139722
|
dmtn
|
dematin actin binding protein |
| chr20_-_4793450 | 1.40 |
ENSDART00000053870
|
galca
|
galactosylceramidase a |
| chr18_+_28106139 | 1.37 |
ENSDART00000089615
|
kiaa1549lb
|
KIAA1549-like b |
| chr15_-_5563551 | 1.35 |
ENSDART00000099520
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr22_+_1796057 | 1.35 |
ENSDART00000170834
|
znf1179
|
zinc finger protein 1179 |
| chr8_+_14792830 | 1.33 |
ENSDART00000139972
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr2_+_3452590 | 1.33 |
ENSDART00000153935
|
cyp8b2
|
cytochrome P450, family 8, subfamily B, polypeptide 2 |
| chr11_+_25112269 | 1.32 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr3_+_15907297 | 1.32 |
ENSDART00000139206
|
mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr6_-_33685325 | 1.32 |
ENSDART00000181883
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr5_+_23118470 | 1.28 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr2_+_20331445 | 1.28 |
ENSDART00000186880
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr6_-_31348999 | 1.26 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr5_+_59252701 | 1.25 |
ENSDART00000144829
|
upk3b
|
uroplakin 3b |
| chr10_+_16501699 | 1.24 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr8_-_25327809 | 1.24 |
ENSDART00000137242
|
eps8l3b
|
EPS8-like 3b |
| chr15_-_1745408 | 1.24 |
ENSDART00000182311
|
stx1a
|
syntaxin 1A (brain) |
| chr18_+_3332999 | 1.24 |
ENSDART00000160857
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr7_-_57933736 | 1.23 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr19_-_41991104 | 1.22 |
ENSDART00000087055
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr7_+_72882446 | 1.21 |
ENSDART00000131113
ENSDART00000174223 |
ADGRL3
|
adhesion G protein-coupled receptor L3 |
| chr1_+_57331813 | 1.21 |
ENSDART00000152440
ENSDART00000062841 |
epn3b
|
epsin 3b |
| chr17_-_50331351 | 1.20 |
ENSDART00000149294
|
otofb
|
otoferlin b |
| chr2_+_36862473 | 1.19 |
ENSDART00000135624
|
si:dkey-193b15.8
|
si:dkey-193b15.8 |
| chr12_+_47280839 | 1.18 |
ENSDART00000187574
|
RYR2
|
ryanodine receptor 2 |
| chr3_+_22905341 | 1.17 |
ENSDART00000111435
|
hdac5
|
histone deacetylase 5 |
| chr17_-_5610514 | 1.17 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr13_-_51973562 | 1.15 |
ENSDART00000166701
|
slc30a10
|
solute carrier family 30, member 10 |
| chr14_+_25464681 | 1.15 |
ENSDART00000067500
ENSDART00000187601 |
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr16_-_29452039 | 1.13 |
ENSDART00000148960
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr7_-_24005268 | 1.13 |
ENSDART00000173608
|
si:dkey-183c6.9
|
si:dkey-183c6.9 |
| chr1_-_45157243 | 1.11 |
ENSDART00000131882
|
mucms1
|
mucin, multiple PTS and SEA group, member 1 |
| chr7_+_42206847 | 1.11 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr3_+_431208 | 1.11 |
ENSDART00000154296
ENSDART00000048733 |
si:ch73-308m11.1
si:dkey-167k11.5
|
si:ch73-308m11.1 si:dkey-167k11.5 |
| chr20_+_27393668 | 1.11 |
ENSDART00000005473
|
tmem179
|
transmembrane protein 179 |
| chr3_-_33029426 | 1.10 |
ENSDART00000181287
|
CT583642.1
|
|
| chr21_-_10773344 | 1.10 |
ENSDART00000063244
|
grp
|
gastrin-releasing peptide |
| chr9_+_33158191 | 1.10 |
ENSDART00000180786
|
dopey2
|
dopey family member 2 |
| chr24_+_32472155 | 1.09 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr24_-_982443 | 1.09 |
ENSDART00000063151
|
napga
|
N-ethylmaleimide-sensitive factor attachment protein, gamma a |
| chr11_+_43401201 | 1.08 |
ENSDART00000190128
|
vipb
|
vasoactive intestinal peptide b |
| chr2_-_24369087 | 1.08 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr23_-_29502287 | 1.08 |
ENSDART00000141075
ENSDART00000053807 |
kif1b
|
kinesin family member 1B |
| chr17_-_51938663 | 1.08 |
ENSDART00000179784
|
ERG28
|
ergosterol biosynthesis 28 homolog |
| chr7_+_11543999 | 1.08 |
ENSDART00000173676
|
il16
|
interleukin 16 |
| chr15_-_36357889 | 1.07 |
ENSDART00000156377
|
si:dkey-23k10.5
|
si:dkey-23k10.5 |
| chr21_+_9576176 | 1.07 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr12_-_10300101 | 1.06 |
ENSDART00000126428
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr3_+_5083407 | 1.03 |
ENSDART00000146883
|
si:ch73-338o16.4
|
si:ch73-338o16.4 |
| chr14_-_30490763 | 1.02 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr11_-_27874116 | 1.01 |
ENSDART00000180579
|
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr4_+_3358383 | 1.01 |
ENSDART00000075320
|
nampta
|
nicotinamide phosphoribosyltransferase a |
| chr13_-_36050303 | 1.01 |
ENSDART00000134955
ENSDART00000139087 |
lgmn
|
legumain |
| chr5_-_63286077 | 1.00 |
ENSDART00000131274
|
tbcelb
|
tubulin folding cofactor E-like b |
| chr18_+_507835 | 1.00 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr11_-_37997419 | 0.99 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr25_+_13191615 | 0.97 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr10_+_23511816 | 0.97 |
ENSDART00000127255
|
zmp:0000000924
|
zmp:0000000924 |
| chr22_-_36774219 | 0.96 |
ENSDART00000056151
ENSDART00000168711 |
acy1
|
aminoacylase 1 |
| chr17_-_24564674 | 0.96 |
ENSDART00000105435
ENSDART00000135086 |
abch1
|
ATP-binding cassette, sub-family H, member 1 |
| chr19_+_17385561 | 0.95 |
ENSDART00000141397
ENSDART00000143913 ENSDART00000133626 |
rpl15
|
ribosomal protein L15 |
| chr18_-_8579907 | 0.94 |
ENSDART00000147284
|
FRMD4A
|
si:ch211-220f12.1 |
| chr24_-_32408404 | 0.94 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr19_-_46088429 | 0.94 |
ENSDART00000161385
|
ptdss1b
|
phosphatidylserine synthase 1b |
| chr25_+_20077225 | 0.93 |
ENSDART00000136543
|
tnni4b.1
|
troponin I4b, tandem duplicate 1 |
| chr4_+_22343093 | 0.92 |
ENSDART00000023588
|
guca1a
|
guanylate cyclase activator 1A |
| chr5_-_21422390 | 0.90 |
ENSDART00000144198
|
tenm1
|
teneurin transmembrane protein 1 |
| chr17_+_33415319 | 0.90 |
ENSDART00000140805
ENSDART00000025501 ENSDART00000146447 |
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr6_+_34512313 | 0.90 |
ENSDART00000102554
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr5_-_41049690 | 0.89 |
ENSDART00000174936
ENSDART00000135030 |
pdzd2
|
PDZ domain containing 2 |
| chr5_-_9625459 | 0.88 |
ENSDART00000143347
|
sh2b3
|
SH2B adaptor protein 3 |
| chr20_+_20637866 | 0.88 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr21_-_2168515 | 0.88 |
ENSDART00000165455
|
BX510922.2
|
|
| chr11_+_30729745 | 0.86 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr19_+_46372115 | 0.86 |
ENSDART00000163935
|
med30
|
mediator complex subunit 30 |
| chr1_+_6817292 | 0.86 |
ENSDART00000145822
ENSDART00000092118 |
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr22_-_20379045 | 0.86 |
ENSDART00000183511
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr7_+_7151832 | 0.85 |
ENSDART00000109485
|
gal3st3
|
galactose-3-O-sulfotransferase 3 |
| chr8_-_554540 | 0.83 |
ENSDART00000163934
|
FO704758.1
|
Danio rerio uncharacterized LOC100329294 (LOC100329294), mRNA. |
| chr20_-_45040916 | 0.83 |
ENSDART00000190001
|
klhl29
|
kelch-like family member 29 |
| chr21_-_7035599 | 0.83 |
ENSDART00000139777
|
si:ch211-93g21.1
|
si:ch211-93g21.1 |
| chr3_-_31833266 | 0.83 |
ENSDART00000084932
ENSDART00000183617 |
map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr12_+_4900981 | 0.83 |
ENSDART00000171507
|
cd79b
|
CD79b molecule, immunoglobulin-associated beta |
| chr7_+_33172066 | 0.82 |
ENSDART00000174013
|
si:ch211-194p6.12
|
si:ch211-194p6.12 |
| chr18_-_15559817 | 0.82 |
ENSDART00000061681
|
si:ch211-245j22.3
|
si:ch211-245j22.3 |
| chr11_+_2506516 | 0.81 |
ENSDART00000130886
ENSDART00000189767 |
NABP2
|
si:ch73-190f16.2 |
| chr9_-_18424844 | 0.80 |
ENSDART00000154351
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr9_+_17423941 | 0.79 |
ENSDART00000112884
ENSDART00000155233 |
kbtbd7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr8_+_51707781 | 0.79 |
ENSDART00000129372
|
klhl22
|
kelch-like family member 22 |
| chr11_-_25734417 | 0.79 |
ENSDART00000103570
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr2_-_36040820 | 0.78 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr19_-_9867001 | 0.78 |
ENSDART00000091695
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr7_+_56889879 | 0.78 |
ENSDART00000039810
|
myo9aa
|
myosin IXAa |
| chr3_-_40976288 | 0.78 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr6_+_4255319 | 0.78 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr3_+_32403758 | 0.77 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr20_+_4793516 | 0.77 |
ENSDART00000053875
|
lgals8a
|
galectin 8a |
| chr5_+_65491390 | 0.77 |
ENSDART00000159921
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr7_+_18100996 | 0.76 |
ENSDART00000055810
|
rab1ba
|
zRAB1B, member RAS oncogene family a |
| chr8_-_4097722 | 0.76 |
ENSDART00000135006
|
cux2b
|
cut-like homeobox 2b |
| chr20_+_14968031 | 0.76 |
ENSDART00000151805
ENSDART00000151448 ENSDART00000063874 ENSDART00000190910 |
vamp4
|
vesicle-associated membrane protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of maff+mafga+mafgb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.7 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.8 | 2.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.4 | 1.8 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.4 | 1.1 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.4 | 2.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.4 | 1.9 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.4 | 3.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.4 | 1.1 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.4 | 1.8 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.3 | 2.9 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.3 | 5.2 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.3 | 1.3 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.3 | 4.8 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 1.5 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.3 | 2.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 2.7 | GO:0098869 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.3 | 1.1 | GO:0070459 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.3 | 1.3 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.3 | 1.3 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.3 | 1.0 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.3 | 1.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 1.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.6 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.2 | 1.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 1.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 0.8 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 1.9 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.2 | 1.4 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 1.2 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.2 | 2.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 0.8 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 0.5 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 0.8 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 1.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 2.8 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 1.5 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 1.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 1.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 2.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 2.0 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 2.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 1.8 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.6 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 1.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.7 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.1 | 0.9 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 5.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 1.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 3.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.6 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.1 | 1.3 | GO:0072028 | nephron morphogenesis(GO:0072028) |
| 0.1 | 1.6 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.7 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 1.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 1.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 7.8 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.1 | 2.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.4 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 1.1 | GO:0050930 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 1.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 4.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.8 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.8 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 1.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 0.5 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 1.0 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 0.6 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.4 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.4 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 1.7 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.1 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.0 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.7 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 3.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.9 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 2.8 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.8 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 1.2 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 2.0 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.6 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 1.2 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 0.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.4 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 3.6 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 1.5 | GO:0006885 | regulation of pH(GO:0006885) regulation of cellular pH(GO:0030641) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.3 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 1.3 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 1.7 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 1.1 | GO:0050922 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.3 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 1.1 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.9 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.6 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.2 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 1.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.1 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 1.2 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 0.4 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 2.4 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.0 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.5 | 2.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.3 | 1.0 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.3 | 4.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 1.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 1.5 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.2 | 0.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 0.7 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.2 | 1.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 2.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.8 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 7.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 4.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 2.0 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 3.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 4.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.8 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 2.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 2.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 2.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 8.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.5 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 8.7 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.6 | 2.9 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.6 | 1.8 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.5 | 4.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.5 | 1.4 | GO:0005183 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.4 | 1.3 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.4 | 2.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 2.7 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 2.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 1.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.3 | 2.8 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 2.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 1.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.3 | 3.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 3.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.2 | 2.0 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 6.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 0.6 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.2 | 2.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 1.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 1.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 1.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.5 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 1.8 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 3.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 2.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.9 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.6 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 2.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.8 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 5.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.6 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 1.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 2.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.9 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 1.6 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 1.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.8 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.7 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.6 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 1.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.2 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.8 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.5 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.0 | 2.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.4 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 2.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 8.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 4.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 2.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.4 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 2.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 3.5 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 2.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.3 | 2.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 2.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 2.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.2 | 2.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 2.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.0 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 2.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 2.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |