Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
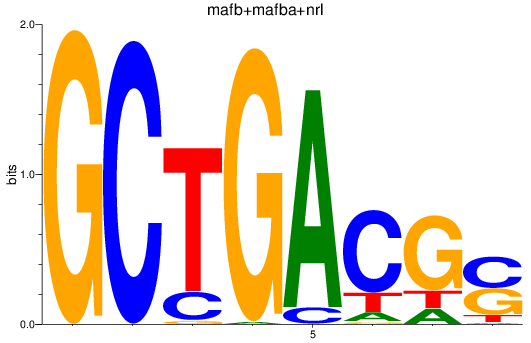
Results for mafb+mafba+nrl
Z-value: 2.39
Transcription factors associated with mafb+mafba+nrl
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafba
|
ENSDARG00000017121 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
|
mafb
|
ENSDARG00000076520 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog b (paralog b) |
|
nrl
|
ENSDARG00000100466 | neural retina leucine zipper |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nrl | dr11_v1_chr20_+_661037_661037 | 0.83 | 1.3e-05 | Click! |
| mafb | dr11_v1_chr25_+_36674715_36674715 | 0.81 | 2.4e-05 | Click! |
| mafba | dr11_v1_chr23_-_3409140_3409140 | -0.28 | 2.5e-01 | Click! |
Activity profile of mafb+mafba+nrl motif
Sorted Z-values of mafb+mafba+nrl motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22318511 | 8.99 |
ENSDART00000129295
|
crygm2d2
|
crystallin, gamma M2d2 |
| chr25_-_3503164 | 6.65 |
ENSDART00000191477
ENSDART00000186345 ENSDART00000180199 |
PRKAR2B
|
si:ch211-272n13.7 |
| chr9_-_22232902 | 6.60 |
ENSDART00000101845
|
crygm2d5
|
crystallin, gamma M2d5 |
| chr1_+_44439661 | 6.59 |
ENSDART00000100309
|
crybb1l2
|
crystallin, beta B1, like 2 |
| chr14_+_49135264 | 5.97 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr3_-_61185746 | 5.91 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr11_-_1291012 | 5.31 |
ENSDART00000158390
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr3_-_61203203 | 5.23 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr8_+_6954984 | 4.80 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr25_-_4482449 | 4.61 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr24_-_41312459 | 4.05 |
ENSDART00000041349
|
crygn2
|
crystallin, gamma N2 |
| chr9_-_22240052 | 4.02 |
ENSDART00000111109
|
crygm2d9
|
crystallin, gamma M2d9 |
| chr8_-_14052349 | 3.93 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr10_-_7858553 | 3.85 |
ENSDART00000182010
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr9_-_22147567 | 3.83 |
ENSDART00000110941
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr17_+_9310259 | 3.77 |
ENSDART00000186158
ENSDART00000190329 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr1_-_44434707 | 3.63 |
ENSDART00000110148
|
cryba1l2
|
crystallin, beta A1, like 2 |
| chr3_-_50443607 | 3.49 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr19_-_5345930 | 3.49 |
ENSDART00000066620
ENSDART00000151398 |
krtt1c19e
|
keratin type 1 c19e |
| chr14_+_904850 | 3.37 |
ENSDART00000161847
|
si:ch73-208h1.1
|
si:ch73-208h1.1 |
| chr23_+_21067684 | 3.37 |
ENSDART00000132066
|
kcnab2b
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 b |
| chr5_-_71722257 | 3.36 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr7_-_26087807 | 3.28 |
ENSDART00000052989
|
ache
|
acetylcholinesterase |
| chr13_+_3954540 | 3.26 |
ENSDART00000092646
|
lrrc73
|
leucine rich repeat containing 73 |
| chr24_-_26283359 | 3.22 |
ENSDART00000128618
|
and1
|
actinodin1 |
| chr6_-_60147517 | 3.18 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr1_-_59287410 | 2.99 |
ENSDART00000158011
ENSDART00000170580 |
col5a3b
|
collagen, type V, alpha 3b |
| chr23_+_2542158 | 2.97 |
ENSDART00000182197
|
LO017835.1
|
|
| chr15_+_2559875 | 2.89 |
ENSDART00000178505
|
SH2B2
|
SH2B adaptor protein 2 |
| chr13_+_24755049 | 2.88 |
ENSDART00000134102
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr8_-_23416362 | 2.84 |
ENSDART00000063005
|
gpr173
|
G protein-coupled receptor 173 |
| chr5_+_36614196 | 2.77 |
ENSDART00000150574
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr8_+_24854600 | 2.75 |
ENSDART00000156570
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr14_-_52521460 | 2.74 |
ENSDART00000172110
|
GPR151
|
G protein-coupled receptor 151 |
| chr15_+_16897554 | 2.73 |
ENSDART00000154679
|
ypel2b
|
yippee-like 2b |
| chr4_+_12113105 | 2.73 |
ENSDART00000182399
|
tmem178b
|
transmembrane protein 178B |
| chr14_+_1365097 | 2.71 |
ENSDART00000083797
ENSDART00000176087 |
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr21_+_41697552 | 2.71 |
ENSDART00000169511
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr15_+_45640906 | 2.67 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr17_-_608857 | 2.63 |
ENSDART00000163431
|
KLHL28
|
kelch like family member 28 |
| chr24_-_31942656 | 2.62 |
ENSDART00000180308
ENSDART00000016879 |
c1ql3a
|
complement component 1, q subcomponent-like 3a |
| chr19_-_5254699 | 2.60 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr7_+_58686860 | 2.60 |
ENSDART00000052332
|
penkb
|
proenkephalin b |
| chr25_-_3503458 | 2.59 |
ENSDART00000173269
|
PRKAR2B
|
si:ch211-272n13.7 |
| chr7_+_72882446 | 2.57 |
ENSDART00000131113
ENSDART00000174223 |
ADGRL3
|
adhesion G protein-coupled receptor L3 |
| chr4_+_26496489 | 2.57 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr5_-_37935607 | 2.54 |
ENSDART00000141233
ENSDART00000084868 ENSDART00000125857 |
scn4bb
|
sodium channel, voltage-gated, type IV, beta b |
| chr23_-_7799184 | 2.52 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr4_-_53370 | 2.51 |
ENSDART00000180254
ENSDART00000186529 |
CU856344.2
|
|
| chr15_+_28096152 | 2.48 |
ENSDART00000100293
ENSDART00000140092 |
crybb1l3
|
crystallin, beta B1, like 3 |
| chr3_-_58455289 | 2.45 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr5_-_70625697 | 2.44 |
ENSDART00000179538
ENSDART00000190540 |
CABZ01075572.1
|
|
| chr22_-_38543630 | 2.44 |
ENSDART00000172029
|
si:ch211-126j24.1
|
si:ch211-126j24.1 |
| chr1_-_21344478 | 2.42 |
ENSDART00000077805
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr9_-_296169 | 2.38 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr24_-_29997145 | 2.37 |
ENSDART00000135094
|
palmdb
|
palmdelphin b |
| chr23_-_7755373 | 2.37 |
ENSDART00000162868
|
PCMTD2 (1 of many)
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr16_+_34528409 | 2.36 |
ENSDART00000144718
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr12_-_3840664 | 2.35 |
ENSDART00000160967
|
taok2b
|
TAO kinase 2b |
| chr1_-_5455498 | 2.31 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr14_+_81919 | 2.30 |
ENSDART00000171444
|
stag3
|
stromal antigen 3 |
| chr1_+_36437585 | 2.28 |
ENSDART00000189182
|
pou4f2
|
POU class 4 homeobox 2 |
| chr19_+_58954 | 2.27 |
ENSDART00000162379
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr11_-_3494964 | 2.26 |
ENSDART00000162369
|
acap3b
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3b |
| chr19_+_233143 | 2.24 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr16_+_1254390 | 2.22 |
ENSDART00000092627
|
ADAMTSL4
|
ADAMTS like 4 |
| chr7_+_53541173 | 2.22 |
ENSDART00000159449
|
gramd2aa
|
GRAM domain containing 2Aa |
| chr17_-_50220228 | 2.21 |
ENSDART00000058707
|
jdp2a
|
Jun dimerization protein 2a |
| chr22_-_600016 | 2.21 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr18_+_660578 | 2.20 |
ENSDART00000161203
|
CELF6
|
si:dkey-205h23.2 |
| chr21_-_45588720 | 2.20 |
ENSDART00000186642
ENSDART00000189531 |
LO018363.2
|
|
| chr19_-_27261102 | 2.19 |
ENSDART00000143919
|
gabbr1b
|
gamma-aminobutyric acid (GABA) B receptor, 1b |
| chr3_+_61185660 | 2.18 |
ENSDART00000167114
|
CR352263.1
|
|
| chr9_+_53707240 | 2.18 |
ENSDART00000171490
|
PCDH8
|
si:ch211-199f5.1 |
| chr1_+_54673846 | 2.18 |
ENSDART00000145018
|
gprc5bb
|
G protein-coupled receptor, class C, group 5, member Bb |
| chr22_+_31821815 | 2.14 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr12_-_7824291 | 2.14 |
ENSDART00000148673
ENSDART00000149453 |
ank3b
|
ankyrin 3b |
| chr13_-_25284379 | 2.13 |
ENSDART00000124167
|
vcla
|
vinculin a |
| chr19_+_49721 | 2.12 |
ENSDART00000160489
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr8_+_53423408 | 2.12 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr22_+_17359346 | 2.11 |
ENSDART00000145434
|
gpr52
|
G protein-coupled receptor 52 |
| chr5_-_33259079 | 2.10 |
ENSDART00000132223
|
ifitm1
|
interferon induced transmembrane protein 1 |
| chr8_+_50983551 | 2.08 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr8_+_53464216 | 2.08 |
ENSDART00000169514
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr17_+_20605013 | 2.08 |
ENSDART00000156878
|
si:ch73-288o11.5
|
si:ch73-288o11.5 |
| chr8_+_1082100 | 2.08 |
ENSDART00000149276
|
lzts3b
|
leucine zipper, putative tumor suppressor family member 3b |
| chr10_+_38775408 | 2.08 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr4_+_19534833 | 2.07 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr9_+_49659114 | 2.06 |
ENSDART00000189643
|
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr22_+_18816662 | 2.06 |
ENSDART00000132476
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr3_+_14317802 | 2.05 |
ENSDART00000162023
|
plppr2a
|
phospholipid phosphatase related 2a |
| chr23_+_44614056 | 2.05 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr17_-_37214196 | 2.05 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr21_+_3244146 | 2.02 |
ENSDART00000127740
|
ctif
|
CBP80/20-dependent translation initiation factor |
| chr13_+_3667230 | 2.01 |
ENSDART00000131553
ENSDART00000189841 ENSDART00000183554 ENSDART00000018737 |
qkib
|
QKI, KH domain containing, RNA binding b |
| chr4_-_73190246 | 2.01 |
ENSDART00000170842
|
LO018260.1
|
|
| chr19_+_37925616 | 2.00 |
ENSDART00000148348
|
nxph1
|
neurexophilin 1 |
| chr21_+_42226113 | 2.00 |
ENSDART00000170362
|
GABRB2 (1 of many)
|
gamma-aminobutyric acid type A receptor beta2 subunit |
| chr21_-_23475361 | 1.99 |
ENSDART00000156658
ENSDART00000157454 |
ncam1a
|
neural cell adhesion molecule 1a |
| chr14_-_2206476 | 1.98 |
ENSDART00000081870
|
pcdh2ab6
|
protocadherin 2 alpha b 6 |
| chr3_-_30118856 | 1.98 |
ENSDART00000109953
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr13_-_25284716 | 1.97 |
ENSDART00000039828
|
vcla
|
vinculin a |
| chr24_-_35707552 | 1.97 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr15_-_434503 | 1.97 |
ENSDART00000122286
|
CABZ01056629.1
|
|
| chr19_-_677713 | 1.96 |
ENSDART00000025146
|
slc6a19a.1
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 1 |
| chr4_+_8797197 | 1.96 |
ENSDART00000158671
|
sult4a1
|
sulfotransferase family 4A, member 1 |
| chr12_-_314899 | 1.94 |
ENSDART00000066579
|
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr24_-_39772045 | 1.94 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr6_-_13188667 | 1.93 |
ENSDART00000191654
|
adam23a
|
ADAM metallopeptidase domain 23a |
| chr18_+_45526585 | 1.93 |
ENSDART00000138511
|
kifc3
|
kinesin family member C3 |
| chr2_+_52232630 | 1.91 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr19_-_1023051 | 1.91 |
ENSDART00000158429
|
tmem42b
|
transmembrane protein 42b |
| chr10_+_20113830 | 1.91 |
ENSDART00000139722
|
dmtn
|
dematin actin binding protein |
| chr15_-_47937204 | 1.89 |
ENSDART00000154705
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr23_+_45845423 | 1.88 |
ENSDART00000183404
|
lmnl3
|
lamin L3 |
| chr20_+_52389858 | 1.88 |
ENSDART00000185863
ENSDART00000166651 |
arhgap39
|
Rho GTPase activating protein 39 |
| chr5_-_22501663 | 1.87 |
ENSDART00000133174
|
si:dkey-27p18.5
|
si:dkey-27p18.5 |
| chr6_-_27139396 | 1.87 |
ENSDART00000055848
|
zgc:103559
|
zgc:103559 |
| chr5_-_12525441 | 1.86 |
ENSDART00000177490
ENSDART00000164190 |
ksr2
|
kinase suppressor of ras 2 |
| chr9_+_25096500 | 1.85 |
ENSDART00000135074
ENSDART00000180436 ENSDART00000108629 |
lrch1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr15_+_24212847 | 1.85 |
ENSDART00000155502
|
sez6b
|
seizure related 6 homolog b |
| chr20_-_37629084 | 1.84 |
ENSDART00000141734
|
hivep2a
|
human immunodeficiency virus type I enhancer binding protein 2a |
| chr13_+_3954715 | 1.84 |
ENSDART00000182477
ENSDART00000192142 ENSDART00000190962 |
lrrc73
|
leucine rich repeat containing 73 |
| chr9_+_30890549 | 1.84 |
ENSDART00000101070
|
dachd
|
dachshund d |
| chr25_-_207214 | 1.80 |
ENSDART00000193448
|
FP236318.3
|
|
| chr7_-_38612230 | 1.79 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr9_-_23217196 | 1.79 |
ENSDART00000083567
|
kif5c
|
kinesin family member 5C |
| chr15_-_2954443 | 1.79 |
ENSDART00000161053
|
zgc:153184
|
zgc:153184 |
| chr5_-_12524996 | 1.77 |
ENSDART00000142258
|
si:ch73-263f13.1
|
si:ch73-263f13.1 |
| chr8_-_43456025 | 1.77 |
ENSDART00000001092
ENSDART00000140618 |
ncor2
|
nuclear receptor corepressor 2 |
| chr9_+_21407987 | 1.77 |
ENSDART00000145627
|
gja3
|
gap junction protein, alpha 3 |
| chr5_-_32092856 | 1.76 |
ENSDART00000086181
ENSDART00000181677 |
cabp7b
|
calcium binding protein 7b |
| chr5_-_28915130 | 1.76 |
ENSDART00000078592
|
npdc1b
|
neural proliferation, differentiation and control, 1b |
| chr22_-_31517300 | 1.76 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr23_+_45845159 | 1.75 |
ENSDART00000023944
|
lmnl3
|
lamin L3 |
| chr1_-_43915423 | 1.75 |
ENSDART00000181915
ENSDART00000113673 |
scpp5
|
secretory calcium-binding phosphoprotein 5 |
| chr5_-_21030934 | 1.74 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr9_+_14023386 | 1.74 |
ENSDART00000140199
ENSDART00000124267 |
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr18_+_3630331 | 1.73 |
ENSDART00000159059
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr23_-_3674443 | 1.73 |
ENSDART00000134830
ENSDART00000057422 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr6_-_15604157 | 1.73 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr3_+_16566361 | 1.72 |
ENSDART00000132869
ENSDART00000080771 |
slc6a16a
|
solute carrier family 6, member 16a |
| chr17_-_43399896 | 1.72 |
ENSDART00000156033
ENSDART00000156418 |
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr2_-_24444838 | 1.72 |
ENSDART00000147885
ENSDART00000164720 |
kcnn1a
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1a |
| chr16_+_10346277 | 1.71 |
ENSDART00000081092
|
si:dkeyp-77h1.4
|
si:dkeyp-77h1.4 |
| chr1_+_49266886 | 1.70 |
ENSDART00000137179
|
caly
|
calcyon neuron-specific vesicular protein |
| chr22_+_2254972 | 1.70 |
ENSDART00000144906
|
znf1157
|
zinc finger protein 1157 |
| chr15_-_47193564 | 1.70 |
ENSDART00000172453
|
LSAMP
|
limbic system-associated membrane protein |
| chr17_+_30450163 | 1.67 |
ENSDART00000104257
|
lpin1
|
lipin 1 |
| chr18_-_85294 | 1.67 |
ENSDART00000044387
|
gabpb1
|
GA binding protein transcription factor, beta subunit 1 |
| chr4_+_68562464 | 1.66 |
ENSDART00000192954
|
BX548011.4
|
|
| chr6_-_57938043 | 1.64 |
ENSDART00000171073
|
tox2
|
TOX high mobility group box family member 2 |
| chr11_+_30161168 | 1.63 |
ENSDART00000157385
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr23_-_45049044 | 1.63 |
ENSDART00000067630
|
SH3GL2 (1 of many)
|
SH3 domain containing GRB2 like 2, endophilin A1 |
| chr23_-_30954738 | 1.62 |
ENSDART00000188996
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr22_+_2143107 | 1.62 |
ENSDART00000172339
ENSDART00000161280 ENSDART00000163671 ENSDART00000172090 ENSDART00000163573 |
znf1164
|
zinc finger protein 1164 |
| chr16_+_10841163 | 1.61 |
ENSDART00000065467
|
dedd1
|
death effector domain-containing 1 |
| chr16_-_43356018 | 1.60 |
ENSDART00000181683
|
FO704821.1
|
|
| chr20_-_53321499 | 1.60 |
ENSDART00000179894
ENSDART00000127427 ENSDART00000084952 |
wasf1
|
WAS protein family, member 1 |
| chr14_-_52542928 | 1.60 |
ENSDART00000075749
|
ppp2r2ba
|
protein phosphatase 2, regulatory subunit B, beta a |
| chr6_+_112579 | 1.60 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr22_+_1137997 | 1.59 |
ENSDART00000158017
ENSDART00000183226 |
si:ch211-103a14.5
|
si:ch211-103a14.5 |
| chr8_+_29267093 | 1.58 |
ENSDART00000077647
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr24_-_31452875 | 1.58 |
ENSDART00000187381
ENSDART00000185128 |
cngb3.2
|
cyclic nucleotide gated channel beta 3, tandem duplicate 2 |
| chr20_-_54462551 | 1.55 |
ENSDART00000171769
ENSDART00000169692 |
evlb
|
Enah/Vasp-like b |
| chr22_+_635813 | 1.55 |
ENSDART00000179067
|
CU856139.1
|
|
| chr14_-_44841335 | 1.55 |
ENSDART00000173011
|
GRXCR1
|
si:dkey-109l4.6 |
| chr9_-_22069364 | 1.54 |
ENSDART00000101938
|
crygm2b
|
crystallin, gamma M2b |
| chr5_+_45976130 | 1.54 |
ENSDART00000175670
|
sv2c
|
synaptic vesicle glycoprotein 2C |
| chr18_-_1228688 | 1.52 |
ENSDART00000064403
|
nptnb
|
neuroplastin b |
| chr19_-_6134802 | 1.51 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr13_-_11644806 | 1.51 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr5_-_68022631 | 1.51 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr23_-_270847 | 1.50 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr8_-_22288258 | 1.50 |
ENSDART00000140978
ENSDART00000100046 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr3_+_20012891 | 1.50 |
ENSDART00000078974
|
tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr1_+_52690448 | 1.49 |
ENSDART00000150326
|
osbp
|
oxysterol binding protein |
| chr14_-_1454045 | 1.49 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr6_+_103361 | 1.49 |
ENSDART00000151899
|
ldlrb
|
low density lipoprotein receptor b |
| chr21_+_382400 | 1.49 |
ENSDART00000190785
ENSDART00000124615 |
rad23b
|
RAD23 homolog B, nucleotide excision repair protein |
| chr14_-_30642819 | 1.49 |
ENSDART00000078154
|
npas4a
|
neuronal PAS domain protein 4a |
| chr18_-_12612699 | 1.49 |
ENSDART00000090335
|
hipk2
|
homeodomain interacting protein kinase 2 |
| chr3_-_53465223 | 1.49 |
ENSDART00000057123
ENSDART00000125515 ENSDART00000143096 |
nr5a5
|
nuclear receptor subfamily 5, group A, member 5 |
| chr10_+_35953068 | 1.48 |
ENSDART00000015279
|
rtn4rl1a
|
reticulon 4 receptor-like 1a |
| chr6_-_58828398 | 1.48 |
ENSDART00000090634
|
kif5ab
|
kinesin family member 5A, b |
| chr18_+_10840071 | 1.48 |
ENSDART00000014496
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr4_+_72798545 | 1.48 |
ENSDART00000181727
|
MYRFL
|
myelin regulatory factor-like |
| chr17_-_43286903 | 1.47 |
ENSDART00000176637
|
EML5
|
si:dkey-1f12.3 |
| chr2_-_56635744 | 1.47 |
ENSDART00000167790
ENSDART00000168160 |
pip5k1cb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b |
| chr6_-_10776254 | 1.45 |
ENSDART00000184008
|
gpr155b
|
G protein-coupled receptor 155b |
| chr12_-_45533034 | 1.45 |
ENSDART00000113004
|
CABZ01068356.1
|
|
| chr6_-_24358732 | 1.45 |
ENSDART00000159595
|
ephx4
|
epoxide hydrolase 4 |
| chr4_-_149334 | 1.45 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr17_-_12385308 | 1.45 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr1_+_29281764 | 1.44 |
ENSDART00000112106
|
fam155a
|
family with sequence similarity 155, member A |
| chr6_-_15604417 | 1.43 |
ENSDART00000157817
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr19_+_56351 | 1.43 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr19_+_823945 | 1.43 |
ENSDART00000142287
|
ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr12_-_11570 | 1.42 |
ENSDART00000186179
|
shisa6
|
shisa family member 6 |
| chr23_+_45579497 | 1.42 |
ENSDART00000110381
|
egr4
|
early growth response 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mafb+mafba+nrl
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.9 | 2.7 | GO:0015824 | proline transport(GO:0015824) |
| 0.8 | 3.3 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.8 | 5.7 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.7 | 4.1 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.7 | 2.0 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 0.6 | 2.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.6 | 1.9 | GO:0000256 | allantoin catabolic process(GO:0000256) purine nucleobase catabolic process(GO:0006145) |
| 0.6 | 1.9 | GO:2000402 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.6 | 3.4 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.6 | 4.4 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.5 | 2.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.5 | 2.7 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.5 | 2.6 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.5 | 1.5 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.5 | 3.4 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.5 | 4.6 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.4 | 1.8 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.4 | 1.3 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) |
| 0.4 | 2.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.4 | 1.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.4 | 1.2 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.4 | 3.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.4 | 1.2 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.3 | 1.4 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.3 | 3.8 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.3 | 1.4 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.3 | 1.3 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.3 | 42.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.3 | 2.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.3 | 1.8 | GO:0043584 | nose development(GO:0043584) |
| 0.3 | 1.5 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.3 | 2.1 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.3 | 1.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.3 | 0.8 | GO:0051583 | synaptic transmission, dopaminergic(GO:0001963) norepinephrine transport(GO:0015874) dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.3 | 1.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 | 0.8 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.3 | 2.2 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.3 | 2.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.3 | 1.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 | 1.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) regulation of glucose import(GO:0046324) |
| 0.3 | 2.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.3 | 2.0 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.3 | 1.3 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.2 | 1.0 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.2 | 1.2 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.2 | 0.7 | GO:0032677 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.2 | 2.7 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.2 | 2.0 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.2 | 0.6 | GO:2000639 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.2 | 1.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.2 | 0.6 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 1.3 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.2 | 2.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 3.1 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 1.4 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 1.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 3.2 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.2 | 0.6 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.2 | 0.6 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.2 | 1.9 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.2 | 1.1 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.2 | 0.9 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 0.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.2 | 1.8 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 | 1.2 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.2 | 2.5 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.2 | 1.5 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.2 | 0.8 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.2 | 0.5 | GO:1902893 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.2 | 1.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 0.5 | GO:0035992 | tendon formation(GO:0035992) |
| 0.2 | 1.1 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.2 | 2.1 | GO:0045162 | neuronal ion channel clustering(GO:0045161) clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.6 | GO:0099565 | excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.1 | 0.6 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.1 | 0.7 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 1.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 1.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.6 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 3.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 1.3 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 3.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.3 | GO:0043901 | negative regulation of multi-organism process(GO:0043901) |
| 0.1 | 1.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.3 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.1 | 0.8 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 2.4 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.1 | 0.7 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.4 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 1.3 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.9 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 0.7 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 0.4 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 2.2 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 1.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 2.0 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.6 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 1.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.7 | GO:0090075 | relaxation of muscle(GO:0090075) |
| 0.1 | 3.0 | GO:0015872 | dopamine transport(GO:0015872) |
| 0.1 | 0.4 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.1 | 1.7 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.1 | 2.7 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 4.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.6 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 1.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 3.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.7 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 1.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.4 | GO:0032979 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 1.0 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 7.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.7 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.6 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 0.6 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 2.1 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.4 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.1 | 0.2 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.1 | 1.6 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.8 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 0.6 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.2 | GO:1903729 | regulation of plasma membrane organization(GO:1903729) |
| 0.1 | 1.1 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.7 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.7 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 0.4 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.1 | 0.4 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.6 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.8 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 0.3 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 1.0 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.1 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.2 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.1 | 0.3 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 1.7 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.1 | 1.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.6 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 1.9 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.1 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 10.4 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 0.7 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.2 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.1 | 1.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.6 | GO:0014074 | response to purine-containing compound(GO:0014074) response to ATP(GO:0033198) response to organophosphorus(GO:0046683) |
| 0.1 | 1.9 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.1 | 0.5 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.4 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 1.1 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 1.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.6 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.5 | GO:0006984 | ER-nucleus signaling pathway(GO:0006984) |
| 0.0 | 0.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 2.2 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.1 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 1.3 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 1.3 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.3 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.4 | GO:1902686 | positive regulation of mitochondrial membrane permeability(GO:0035794) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.4 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 1.0 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 1.0 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 4.1 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 1.0 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.6 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 1.7 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 2.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 1.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.7 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 1.3 | GO:0046058 | cAMP biosynthetic process(GO:0006171) cAMP metabolic process(GO:0046058) |
| 0.0 | 2.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 3.1 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.4 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 1.4 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 1.2 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 4.0 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.7 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.5 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.5 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.2 | GO:0033151 | V(D)J recombination(GO:0033151) |
| 0.0 | 1.0 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.3 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 1.2 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 5.5 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.4 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 1.2 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) peptidyl-tyrosine modification(GO:0018212) |
| 0.0 | 3.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 2.7 | GO:0033674 | positive regulation of kinase activity(GO:0033674) |
| 0.0 | 2.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.2 | GO:0009190 | cyclic nucleotide biosynthetic process(GO:0009190) |
| 0.0 | 0.4 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 1.5 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.1 | GO:1902623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.6 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.5 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.8 | GO:0099531 | presynaptic process involved in chemical synaptic transmission(GO:0099531) |
| 0.0 | 0.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.0 | 0.6 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 1.1 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.3 | GO:0050868 | negative regulation of homotypic cell-cell adhesion(GO:0034111) negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.4 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 0.3 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.4 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.5 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.1 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 2.4 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.4 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.4 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 1.9 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.4 | 2.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.4 | 2.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.3 | 1.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 0.9 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.3 | 1.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.3 | 2.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.3 | 2.2 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.3 | 3.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 2.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 0.9 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 1.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.2 | 0.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 3.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 3.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 0.9 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 8.3 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.1 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 3.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 1.3 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.1 | 3.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 2.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 1.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 2.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 10.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.9 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.8 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 3.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 1.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 2.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 0.4 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 1.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.8 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 4.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 0.5 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 0.7 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 3.1 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 3.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 4.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.2 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.1 | 4.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 4.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 1.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 5.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.8 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.3 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 2.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 6.3 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 3.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.0 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 4.5 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 2.1 | GO:0099503 | secretory vesicle(GO:0099503) |
| 0.0 | 0.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 3.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.5 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 5.5 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.6 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 1.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.3 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.7 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.1 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 20.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.2 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.9 | 2.7 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) |
| 0.6 | 4.4 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.5 | 3.3 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.5 | 43.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.5 | 1.9 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.4 | 2.1 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 1.2 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.4 | 1.1 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.4 | 1.8 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.3 | 3.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 1.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.3 | 2.6 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.3 | 1.0 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.3 | 9.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.3 | 0.8 | GO:0005330 | dopamine transmembrane transporter activity(GO:0005329) dopamine:sodium symporter activity(GO:0005330) norepinephrine transmembrane transporter activity(GO:0005333) norepinephrine:sodium symporter activity(GO:0005334) |
| 0.3 | 2.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 3.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 2.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 0.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 3.2 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.2 | 0.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 0.9 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.2 | 2.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 1.9 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.2 | 0.6 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.2 | 0.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 1.0 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.2 | 1.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 1.6 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 0.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 1.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 2.5 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 1.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 0.7 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.2 | 5.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 0.5 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.2 | 9.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 0.5 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.2 | 0.5 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) inositol trisphosphate kinase activity(GO:0051766) |
| 0.2 | 2.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.2 | 1.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 1.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 1.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 1.0 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 1.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 4.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.8 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.7 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.4 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.1 | 0.8 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 1.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.3 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.1 | 3.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 3.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.5 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 1.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.2 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.9 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.8 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 3.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 4.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 1.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 1.9 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 1.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.5 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 1.9 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 3.2 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.1 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 2.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.9 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.9 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 1.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 1.2 | GO:0022884 | macromolecule transmembrane transporter activity(GO:0022884) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 1.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 2.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.1 | 1.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 11.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 0.2 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.1 | 0.9 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 3.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 7.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.0 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 4.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 2.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 2.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.5 | GO:0031420 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.6 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.2 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 1.3 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 1.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 2.2 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 1.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.9 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 2.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.5 | GO:0016502 | G-protein coupled nucleotide receptor activity(GO:0001608) purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) purinergic receptor activity(GO:0035586) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 9.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 1.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 4.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 2.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.4 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 22.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 2.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.7 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 1.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.7 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 1.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0019957 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 5.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.4 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 0.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 2.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 1.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 2.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.4 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.6 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 2.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.5 | 2.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 6.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 4.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.3 | 1.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 3.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 1.8 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.2 | 4.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 2.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 0.7 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.2 | 2.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 2.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 1.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 1.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.6 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 0.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 2.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.9 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 2.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.3 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |