Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
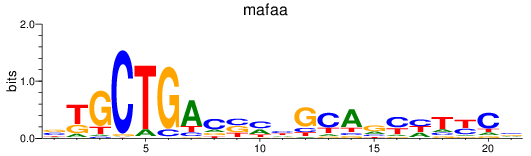
Results for mafaa
Z-value: 0.50
Transcription factors associated with mafaa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafaa
|
ENSDARG00000044155 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mafaa | dr11_v1_chr6_-_32703317_32703317 | 0.01 | 9.8e-01 | Click! |
Activity profile of mafaa motif
Sorted Z-values of mafaa motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_41312459 | 0.99 |
ENSDART00000041349
|
crygn2
|
crystallin, gamma N2 |
| chr16_+_1353894 | 0.80 |
ENSDART00000148426
|
celf3b
|
cugbp, Elav-like family member 3b |
| chr22_-_15280638 | 0.70 |
ENSDART00000063008
|
mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr23_-_21453614 | 0.68 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr1_+_59090972 | 0.67 |
ENSDART00000171497
|
mfap4
|
microfibril associated protein 4 |
| chr1_+_59090743 | 0.66 |
ENSDART00000100199
|
mfap4
|
microfibril associated protein 4 |
| chr23_+_3538463 | 0.64 |
ENSDART00000172758
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr12_-_47558276 | 0.63 |
ENSDART00000160260
|
grem2b
|
gremlin 2, DAN family BMP antagonist b |
| chr23_+_21459263 | 0.59 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr11_+_12719944 | 0.58 |
ENSDART00000054837
ENSDART00000144954 |
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr10_+_21780250 | 0.55 |
ENSDART00000183782
|
pcdh1g9
|
protocadherin 1 gamma 9 |
| chr1_+_59090583 | 0.55 |
ENSDART00000150658
|
mfap4
|
microfibril associated protein 4 |
| chr25_-_27842654 | 0.53 |
ENSDART00000154852
ENSDART00000156906 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr19_-_28788466 | 0.52 |
ENSDART00000151793
|
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr11_-_36263886 | 0.52 |
ENSDART00000140397
|
nfya
|
nuclear transcription factor Y, alpha |
| chr1_-_625875 | 0.38 |
ENSDART00000167331
|
appa
|
amyloid beta (A4) precursor protein a |
| chr1_-_26045007 | 0.36 |
ENSDART00000129259
|
gpank1
|
G patch domain and ankyrin repeats 1 |
| chr25_+_3217419 | 0.36 |
ENSDART00000104859
|
rccd1
|
RCC1 domain containing 1 |
| chr14_-_17588345 | 0.35 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr6_-_39313027 | 0.34 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr20_+_54323218 | 0.33 |
ENSDART00000191096
|
CABZ01077555.1
|
|
| chr13_-_28272299 | 0.33 |
ENSDART00000006393
|
tlx1
|
T cell leukemia homeobox 1 |
| chr19_+_29798064 | 0.30 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr8_-_2591654 | 0.30 |
ENSDART00000049109
|
seta
|
SET nuclear proto-oncogene a |
| chr6_-_33913184 | 0.29 |
ENSDART00000146373
|
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr22_-_775776 | 0.28 |
ENSDART00000149749
|
rbbp5
|
retinoblastoma binding protein 5 |
| chr19_-_3777217 | 0.27 |
ENSDART00000160510
|
si:dkey-206d17.15
|
si:dkey-206d17.15 |
| chr8_+_50534948 | 0.26 |
ENSDART00000174435
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr14_-_30490763 | 0.25 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr16_-_27676123 | 0.25 |
ENSDART00000180804
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr12_+_30789611 | 0.25 |
ENSDART00000181501
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr22_+_635813 | 0.25 |
ENSDART00000179067
|
CU856139.1
|
|
| chr16_-_46567344 | 0.25 |
ENSDART00000127721
|
si:dkey-152b24.7
|
si:dkey-152b24.7 |
| chr18_-_48745517 | 0.25 |
ENSDART00000097259
|
st3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr20_-_23440955 | 0.24 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr23_+_42338325 | 0.23 |
ENSDART00000169660
|
cyp2aa7
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 |
| chr4_+_19700308 | 0.22 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr25_-_3217115 | 0.22 |
ENSDART00000032390
|
gtf2h1
|
general transcription factor IIH, polypeptide 1 |
| chr17_-_48915427 | 0.20 |
ENSDART00000054781
|
lgals8b
|
galectin 8b |
| chr5_-_35137365 | 0.20 |
ENSDART00000141218
|
fcho2
|
FCH domain only 2 |
| chr20_-_39391833 | 0.20 |
ENSDART00000135149
|
si:dkey-217m5.8
|
si:dkey-217m5.8 |
| chr13_+_4671698 | 0.20 |
ENSDART00000164617
ENSDART00000128494 ENSDART00000165776 |
pla2g12b
|
phospholipase A2, group XIIB |
| chr13_-_50463938 | 0.20 |
ENSDART00000083857
|
ccnj
|
cyclin J |
| chr21_-_13225402 | 0.19 |
ENSDART00000080347
|
wdr34
|
WD repeat domain 34 |
| chr8_-_3336273 | 0.18 |
ENSDART00000081160
|
zgc:173737
|
zgc:173737 |
| chr22_+_10606863 | 0.18 |
ENSDART00000147975
|
rad54l2
|
RAD54 like 2 |
| chr15_-_17138640 | 0.17 |
ENSDART00000080777
|
mrpl28
|
mitochondrial ribosomal protein L28 |
| chr7_+_6969909 | 0.17 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr22_+_21305682 | 0.17 |
ENSDART00000023521
|
odf3l2
|
outer dense fiber of sperm tails 3-like 2 |
| chr21_-_17037907 | 0.16 |
ENSDART00000101263
|
ube2g1b
|
ubiquitin-conjugating enzyme E2G 1b (UBC7 homolog, yeast) |
| chr13_+_6295173 | 0.16 |
ENSDART00000163855
|
angpt2a
|
angiopoietin 2a |
| chr21_-_1642739 | 0.16 |
ENSDART00000151118
|
zgc:152948
|
zgc:152948 |
| chr20_-_39219537 | 0.15 |
ENSDART00000005764
|
cyp39a1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr24_+_6353394 | 0.15 |
ENSDART00000165118
|
CR352329.1
|
|
| chr7_+_41421998 | 0.14 |
ENSDART00000083967
|
chpf2
|
chondroitin polymerizing factor 2 |
| chr3_+_8826905 | 0.14 |
ENSDART00000167474
ENSDART00000168088 |
si:dkeyp-30d6.2
|
si:dkeyp-30d6.2 |
| chr8_-_14052349 | 0.13 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr5_-_33230794 | 0.13 |
ENSDART00000144273
|
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr2_+_38161318 | 0.13 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr25_-_21156678 | 0.13 |
ENSDART00000156257
|
wnk1a
|
WNK lysine deficient protein kinase 1a |
| chr12_+_12789853 | 0.10 |
ENSDART00000064839
|
grid1b
|
glutamate receptor, ionotropic, delta 1b |
| chr3_-_10677890 | 0.10 |
ENSDART00000155382
ENSDART00000171319 |
si:ch1073-144j5.2
|
si:ch1073-144j5.2 |
| chr24_+_37709191 | 0.10 |
ENSDART00000066558
|
decr2
|
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr15_+_7086327 | 0.10 |
ENSDART00000114560
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr10_-_2788668 | 0.10 |
ENSDART00000131749
ENSDART00000124356 ENSDART00000085031 |
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr17_-_20202725 | 0.10 |
ENSDART00000133650
|
ecd
|
ecdysoneless homolog (Drosophila) |
| chr4_-_72468168 | 0.09 |
ENSDART00000182995
ENSDART00000174067 |
CR788316.1
|
|
| chr1_-_16676177 | 0.09 |
ENSDART00000159038
|
frg1
|
FSHD region gene 1 |
| chr2_+_36828243 | 0.09 |
ENSDART00000135754
ENSDART00000140474 |
nrd1a
|
nardilysin a (N-arginine dibasic convertase) |
| chr12_-_19119176 | 0.08 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr21_+_19347655 | 0.08 |
ENSDART00000093155
|
hpse
|
heparanase |
| chr8_+_52479026 | 0.08 |
ENSDART00000162885
|
FO704661.1
|
Danio rerio gamma-glutamyl hydrolase (LOC553228), mRNA. |
| chr9_-_1639884 | 0.08 |
ENSDART00000062850
ENSDART00000150947 |
agps
|
alkylglycerone phosphate synthase |
| chr7_-_40578733 | 0.07 |
ENSDART00000173926
ENSDART00000010035 |
dnajb6b
|
DnaJ (Hsp40) homolog, subfamily B, member 6b |
| chr10_-_57270 | 0.06 |
ENSDART00000058411
|
hlcs
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
| chr5_-_40510397 | 0.06 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr12_-_11649690 | 0.06 |
ENSDART00000149713
|
btbd16
|
BTB (POZ) domain containing 16 |
| chr3_+_26322596 | 0.06 |
ENSDART00000172700
|
si:ch211-156b7.5
|
si:ch211-156b7.5 |
| chr25_-_24240797 | 0.06 |
ENSDART00000132790
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr25_+_459901 | 0.05 |
ENSDART00000154895
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr22_+_5574952 | 0.05 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr5_-_23795688 | 0.05 |
ENSDART00000099084
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr24_+_80653 | 0.04 |
ENSDART00000158473
ENSDART00000129135 |
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr6_-_13709591 | 0.04 |
ENSDART00000151771
|
chpfb
|
chondroitin polymerizing factor b |
| chr7_-_7493758 | 0.04 |
ENSDART00000036703
|
pfdn2
|
prefoldin subunit 2 |
| chr7_+_27603211 | 0.04 |
ENSDART00000148782
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr22_+_12477996 | 0.04 |
ENSDART00000177704
|
CR847870.3
|
|
| chr2_-_37896965 | 0.03 |
ENSDART00000129852
|
hbl1
|
hexose-binding lectin 1 |
| chr19_-_42566527 | 0.03 |
ENSDART00000147818
ENSDART00000142135 |
si:dkey-228a15.1
|
si:dkey-228a15.1 |
| chr6_-_25371196 | 0.03 |
ENSDART00000187291
|
PKN2 (1 of many)
|
zgc:153916 |
| chr4_-_50926767 | 0.03 |
ENSDART00000183430
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr15_-_45110011 | 0.02 |
ENSDART00000182047
ENSDART00000188662 |
CABZ01072607.1
|
|
| chr23_+_42652877 | 0.02 |
ENSDART00000035654
|
cyp2ae1
|
cytochrome P450, family 2, subfamily AE, polypeptide 1 |
| chr4_-_8030583 | 0.02 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr20_-_22476255 | 0.02 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr16_+_42465518 | 0.02 |
ENSDART00000058699
|
si:ch211-215k15.4
|
si:ch211-215k15.4 |
| chr8_-_21071476 | 0.01 |
ENSDART00000184184
ENSDART00000100288 |
zgc:112962
|
zgc:112962 |
| chr6_-_131401 | 0.00 |
ENSDART00000151251
|
LRRC8E
|
si:zfos-323e3.4 |
| chr4_+_9911534 | 0.00 |
ENSDART00000109452
|
sbf1
|
SET binding factor 1 |
| chr18_-_26126258 | 0.00 |
ENSDART00000145875
|
ANKRD34C
|
ankyrin repeat domain 34C |
Network of associatons between targets according to the STRING database.
First level regulatory network of mafaa
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 1.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.3 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.5 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) arachidonate transport(GO:1903963) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.0 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 1.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |