Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
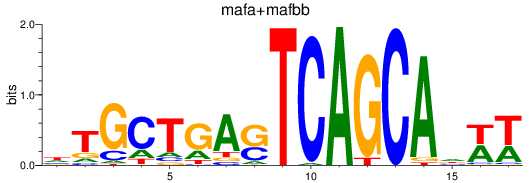
Results for mafa+mafbb
Z-value: 0.80
Transcription factors associated with mafa+mafbb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafa
|
ENSDARG00000015890 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
|
mafbb
|
ENSDARG00000070542 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Bb |
|
mafbb
|
ENSDARG00000099120 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Bb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mafbb | dr11_v1_chr11_-_25384213_25384213 | -0.50 | 3.0e-02 | Click! |
| mafa | dr11_v1_chr18_+_29403017_29403017 | -0.18 | 4.5e-01 | Click! |
Activity profile of mafa+mafbb motif
Sorted Z-values of mafa+mafbb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_13783604 | 5.91 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr5_-_30615901 | 4.00 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr5_-_67471375 | 3.84 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr25_+_31277415 | 3.60 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr9_-_22318511 | 3.11 |
ENSDART00000129295
|
crygm2d2
|
crystallin, gamma M2d2 |
| chr7_+_39386982 | 2.59 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr9_-_22076368 | 2.13 |
ENSDART00000128486
|
crygm2a
|
crystallin, gamma M2a |
| chr21_+_45496442 | 1.94 |
ENSDART00000164880
|
si:dkey-223p19.1
|
si:dkey-223p19.1 |
| chr20_-_14054083 | 1.91 |
ENSDART00000009549
|
rhag
|
Rh associated glycoprotein |
| chr13_+_36770738 | 1.84 |
ENSDART00000146696
|
atl1
|
atlastin GTPase 1 |
| chr18_+_22302635 | 1.60 |
ENSDART00000141051
|
carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr6_-_40744720 | 1.57 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr24_+_3307857 | 1.55 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr17_-_19345521 | 1.51 |
ENSDART00000082085
|
gsc
|
goosecoid |
| chr15_+_45640906 | 1.48 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr23_-_29376859 | 1.48 |
ENSDART00000146411
|
sst6
|
somatostatin 6 |
| chr4_-_16341801 | 1.45 |
ENSDART00000140190
|
kera
|
keratocan |
| chr3_-_18030938 | 1.44 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr7_-_19043401 | 1.27 |
ENSDART00000158796
|
fcer1g
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
| chr10_-_39011514 | 1.23 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr14_+_34547554 | 1.20 |
ENSDART00000074819
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr2_-_9748039 | 1.17 |
ENSDART00000134870
|
si:ch1073-170o4.1
|
si:ch1073-170o4.1 |
| chr8_-_14052349 | 1.12 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr16_-_12319822 | 1.11 |
ENSDART00000127453
ENSDART00000184526 |
trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr23_+_24124684 | 1.06 |
ENSDART00000144478
|
si:dkey-21o19.2
|
si:dkey-21o19.2 |
| chr10_+_31222433 | 1.06 |
ENSDART00000185080
|
tmem218
|
transmembrane protein 218 |
| chr16_-_44349845 | 1.04 |
ENSDART00000170932
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr23_-_20764227 | 1.01 |
ENSDART00000089750
|
znf362b
|
zinc finger protein 362b |
| chr21_-_28523548 | 1.00 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr16_+_47207691 | 1.00 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr16_+_18974064 | 0.99 |
ENSDART00000079248
|
slc6a19b
|
solute carrier family 6 (neutral amino acid transporter), member 19b |
| chr11_-_33960318 | 0.99 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr18_-_8885792 | 0.96 |
ENSDART00000143619
|
si:dkey-95h12.1
|
si:dkey-95h12.1 |
| chr22_-_16416882 | 0.95 |
ENSDART00000062749
|
cts12
|
cathepsin 12 |
| chr10_+_466926 | 0.95 |
ENSDART00000145220
|
arvcfa
|
ARVCF, delta catenin family member a |
| chr20_+_18661624 | 0.95 |
ENSDART00000152136
ENSDART00000126959 |
tnfaip2a
|
tumor necrosis factor, alpha-induced protein 2a |
| chr20_-_29420713 | 0.93 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr22_-_21150845 | 0.92 |
ENSDART00000027345
|
tmem59l
|
transmembrane protein 59-like |
| chr12_+_18556929 | 0.92 |
ENSDART00000191277
|
CT009714.1
|
|
| chr11_+_43401592 | 0.90 |
ENSDART00000112468
|
vipb
|
vasoactive intestinal peptide b |
| chr7_+_35068036 | 0.89 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr13_+_2908764 | 0.88 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr23_-_19500559 | 0.88 |
ENSDART00000177414
ENSDART00000145898 |
asb14b
|
ankyrin repeat and SOCS box containing 14b |
| chr3_-_21061931 | 0.87 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr19_+_31771270 | 0.84 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr10_-_29120515 | 0.80 |
ENSDART00000162016
ENSDART00000149140 |
zpld1a
|
zona pellucida-like domain containing 1a |
| chr11_+_43401201 | 0.80 |
ENSDART00000190128
|
vipb
|
vasoactive intestinal peptide b |
| chr7_+_50053339 | 0.80 |
ENSDART00000174308
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr10_+_26747755 | 0.77 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr6_-_35439406 | 0.77 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr5_-_37116265 | 0.76 |
ENSDART00000057613
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr1_-_26131088 | 0.72 |
ENSDART00000193973
ENSDART00000054209 |
cdkn2a/b
|
cyclin-dependent kinase inhibitor 2A/B (p15, inhibits CDK4) |
| chr1_-_12278522 | 0.72 |
ENSDART00000142122
ENSDART00000003825 |
cplx2l
|
complexin 2, like |
| chr23_-_44494401 | 0.67 |
ENSDART00000114640
ENSDART00000148532 |
actl6b
|
actin-like 6B |
| chr2_-_9059955 | 0.67 |
ENSDART00000022768
|
ak5
|
adenylate kinase 5 |
| chr1_+_8601935 | 0.66 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr5_-_37935607 | 0.65 |
ENSDART00000141233
ENSDART00000084868 ENSDART00000125857 |
scn4bb
|
sodium channel, voltage-gated, type IV, beta b |
| chr5_+_9377005 | 0.63 |
ENSDART00000124924
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr3_+_14463941 | 0.62 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr25_+_19955598 | 0.61 |
ENSDART00000091547
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr24_+_41940299 | 0.60 |
ENSDART00000022349
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr1_-_45888608 | 0.60 |
ENSDART00000139219
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr21_+_21374277 | 0.59 |
ENSDART00000079431
|
rtn2b
|
reticulon 2b |
| chr14_-_4076480 | 0.58 |
ENSDART00000059231
|
enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr22_-_263117 | 0.58 |
ENSDART00000158134
|
zgc:66156
|
zgc:66156 |
| chr11_+_45299447 | 0.56 |
ENSDART00000172999
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr9_-_2573121 | 0.54 |
ENSDART00000181340
|
scrn3
|
secernin 3 |
| chr18_+_10926197 | 0.53 |
ENSDART00000192387
|
ttc38
|
tetratricopeptide repeat domain 38 |
| chr9_-_30247961 | 0.50 |
ENSDART00000131519
|
si:dkey-100n23.3
|
si:dkey-100n23.3 |
| chr7_+_23875269 | 0.49 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr21_-_11820379 | 0.49 |
ENSDART00000126640
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr7_+_61764040 | 0.49 |
ENSDART00000056745
|
acox3
|
acyl-CoA oxidase 3, pristanoyl |
| chr23_-_35082494 | 0.47 |
ENSDART00000189809
|
BX294434.1
|
|
| chr16_-_49781481 | 0.46 |
ENSDART00000127513
|
znf385d
|
zinc finger protein 385D |
| chr10_+_10801564 | 0.46 |
ENSDART00000027026
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr18_-_45617146 | 0.46 |
ENSDART00000146543
|
wt1b
|
wilms tumor 1b |
| chr23_-_36823932 | 0.46 |
ENSDART00000142305
|
hipk1a
|
homeodomain interacting protein kinase 1a |
| chr20_-_20613584 | 0.46 |
ENSDART00000140991
|
si:ch211-253p18.2
|
si:ch211-253p18.2 |
| chr2_-_4496358 | 0.45 |
ENSDART00000158216
|
adipoqb
|
adiponectin, C1Q and collagen domain containing, b |
| chr17_+_51262556 | 0.45 |
ENSDART00000186748
ENSDART00000181606 ENSDART00000063738 ENSDART00000189066 |
eipr1
|
EARP complex and GARP complex interacting protein 1 |
| chr14_-_49859747 | 0.43 |
ENSDART00000169456
ENSDART00000164967 |
nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr9_+_32930622 | 0.42 |
ENSDART00000100928
|
cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr20_-_39219537 | 0.39 |
ENSDART00000005764
|
cyp39a1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr20_-_8304271 | 0.38 |
ENSDART00000179057
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr13_-_20381485 | 0.37 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr12_-_3940768 | 0.37 |
ENSDART00000134292
|
zgc:92040
|
zgc:92040 |
| chr16_+_43368572 | 0.36 |
ENSDART00000032778
ENSDART00000193897 |
rnf144b
|
ring finger protein 144B |
| chr10_+_10801719 | 0.36 |
ENSDART00000193648
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr11_+_40544269 | 0.35 |
ENSDART00000175424
ENSDART00000184673 |
mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr4_-_67909331 | 0.34 |
ENSDART00000191908
|
si:ch211-66c13.1
|
si:ch211-66c13.1 |
| chr25_+_336503 | 0.34 |
ENSDART00000160395
|
CU929262.1
|
|
| chr12_+_16967715 | 0.33 |
ENSDART00000138174
|
slc16a12b
|
solute carrier family 16, member 12b |
| chr22_-_38516922 | 0.33 |
ENSDART00000151257
ENSDART00000151785 |
klc4
|
kinesin light chain 4 |
| chr9_+_14291932 | 0.32 |
ENSDART00000183119
|
BX511082.1
|
|
| chr12_-_44043285 | 0.30 |
ENSDART00000163074
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr5_+_38679623 | 0.30 |
ENSDART00000131879
ENSDART00000170010 |
si:dkey-58f10.11
si:dkey-58f10.10
|
si:dkey-58f10.11 si:dkey-58f10.10 |
| chr19_+_7567763 | 0.30 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr9_+_13985567 | 0.30 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr17_-_51262430 | 0.30 |
ENSDART00000163222
|
trappc12
|
trafficking protein particle complex 12 |
| chr14_+_3507326 | 0.29 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr4_-_58964138 | 0.29 |
ENSDART00000150259
|
si:ch211-64i20.3
|
si:ch211-64i20.3 |
| chr25_-_13558172 | 0.28 |
ENSDART00000139970
ENSDART00000133789 ENSDART00000144426 |
ano10b
|
anoctamin 10b |
| chr8_-_13177536 | 0.27 |
ENSDART00000100992
|
zgc:194990
|
zgc:194990 |
| chr17_-_37312922 | 0.26 |
ENSDART00000153555
|
si:ch211-180n24.3
|
si:ch211-180n24.3 |
| chr21_-_5393125 | 0.26 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr14_+_146857 | 0.26 |
ENSDART00000122521
|
CABZ01088229.1
|
|
| chr12_+_46608361 | 0.25 |
ENSDART00000189164
|
FADS6
|
fatty acid desaturase 6 |
| chr23_+_4253957 | 0.25 |
ENSDART00000008761
|
arl6ip5b
|
ADP-ribosylation factor-like 6 interacting protein 5b |
| chr1_+_50613868 | 0.24 |
ENSDART00000111114
|
HERC5
|
si:ch73-190m4.1 |
| chr20_+_46385907 | 0.23 |
ENSDART00000060710
|
adgrg11
|
adhesion G protein-coupled receptor G11 |
| chr7_+_34790768 | 0.23 |
ENSDART00000075116
|
si:dkey-148a17.6
|
si:dkey-148a17.6 |
| chr22_-_10354381 | 0.23 |
ENSDART00000092050
|
stab1
|
stabilin 1 |
| chr8_+_31435452 | 0.21 |
ENSDART00000145282
|
selenop
|
selenoprotein P |
| chr5_+_38684651 | 0.21 |
ENSDART00000137852
|
si:dkey-58f10.10
|
si:dkey-58f10.10 |
| chr4_+_42878556 | 0.20 |
ENSDART00000171047
|
si:zfos-451d2.2
|
si:zfos-451d2.2 |
| chr1_+_54124209 | 0.20 |
ENSDART00000187730
|
LO017722.1
|
|
| chr13_-_37642890 | 0.20 |
ENSDART00000146483
ENSDART00000136071 ENSDART00000111786 |
si:dkey-188i13.9
|
si:dkey-188i13.9 |
| chr5_-_31035198 | 0.20 |
ENSDART00000086534
|
cyb5d2
|
cytochrome b5 domain containing 2 |
| chr22_+_28818291 | 0.19 |
ENSDART00000136032
|
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr22_-_25469751 | 0.19 |
ENSDART00000171670
|
CR769772.4
|
|
| chr14_-_24001825 | 0.18 |
ENSDART00000130704
|
il4
|
interleukin 4 |
| chr16_-_51299061 | 0.18 |
ENSDART00000148677
|
serpinb1l4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 4 |
| chr7_+_31319876 | 0.17 |
ENSDART00000187611
|
fam189a1
|
family with sequence similarity 189, member A1 |
| chr1_+_16600690 | 0.17 |
ENSDART00000162164
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr23_-_31428763 | 0.16 |
ENSDART00000053545
|
zgc:153284
|
zgc:153284 |
| chr23_-_26077038 | 0.16 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr20_+_38285671 | 0.16 |
ENSDART00000061432
|
ccl38a.4
|
chemokine (C-C motif) ligand 38, duplicate 4 |
| chr1_-_7582859 | 0.16 |
ENSDART00000110696
|
mxb
|
myxovirus (influenza) resistance B |
| chr7_+_29080684 | 0.16 |
ENSDART00000173709
ENSDART00000173576 |
acd
|
ACD, shelterin complex subunit and telomerase recruitment factor |
| chr17_+_23938283 | 0.16 |
ENSDART00000184391
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr6_-_41138854 | 0.15 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr21_+_45659525 | 0.15 |
ENSDART00000160391
|
slc22a5
|
solute carrier family 22 (organic cation/carnitine transporter), member 5 |
| chr7_+_39011355 | 0.15 |
ENSDART00000173855
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr23_+_7518294 | 0.15 |
ENSDART00000081536
|
hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr10_-_37337579 | 0.14 |
ENSDART00000147705
|
omga
|
oligodendrocyte myelin glycoprotein a |
| chr18_+_32615075 | 0.14 |
ENSDART00000166937
|
CABZ01012885.1
|
|
| chr8_-_22698651 | 0.14 |
ENSDART00000181411
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr3_+_57788761 | 0.14 |
ENSDART00000149741
|
si:ch73-362i18.1
|
si:ch73-362i18.1 |
| chr17_-_31044803 | 0.14 |
ENSDART00000185941
ENSDART00000152016 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr1_+_7189856 | 0.13 |
ENSDART00000092114
|
si:ch73-383l1.1
|
si:ch73-383l1.1 |
| chr20_-_20932760 | 0.13 |
ENSDART00000152415
ENSDART00000039907 |
btbd6b
|
BTB (POZ) domain containing 6b |
| chr17_-_2817035 | 0.13 |
ENSDART00000152002
|
gpr65
|
G protein-coupled receptor 65 |
| chr20_-_23852174 | 0.12 |
ENSDART00000122414
|
si:dkey-15j16.6
|
si:dkey-15j16.6 |
| chr16_+_7662609 | 0.12 |
ENSDART00000184895
ENSDART00000149404 ENSDART00000081418 ENSDART00000081422 |
bves
|
blood vessel epicardial substance |
| chr19_+_23308419 | 0.11 |
ENSDART00000141558
|
irgf2
|
immunity-related GTPase family, f2 |
| chr14_-_2203124 | 0.11 |
ENSDART00000124171
|
si:dkeyp-115d7.2
|
si:dkeyp-115d7.2 |
| chr17_+_6217704 | 0.11 |
ENSDART00000129100
|
BX000363.1
|
|
| chr10_+_8527196 | 0.10 |
ENSDART00000141147
|
si:ch211-193e13.5
|
si:ch211-193e13.5 |
| chr12_-_44019561 | 0.10 |
ENSDART00000165900
|
si:dkey-31i7.1
|
si:dkey-31i7.1 |
| chr4_+_65057234 | 0.10 |
ENSDART00000162985
|
si:dkey-14o6.2
|
si:dkey-14o6.2 |
| chr6_+_40723554 | 0.10 |
ENSDART00000103833
|
slc26a6l
|
solute carrier family 26, member 6, like |
| chr9_-_8670158 | 0.10 |
ENSDART00000077296
|
col4a1
|
collagen, type IV, alpha 1 |
| chr19_+_170705 | 0.09 |
ENSDART00000053622
|
rnf139
|
ring finger protein 139 |
| chr4_+_9609905 | 0.09 |
ENSDART00000142284
ENSDART00000150687 |
meig1
|
meiosis/spermiogenesis associated 1 |
| chr3_+_23063718 | 0.09 |
ENSDART00000140225
ENSDART00000184431 |
b4galnt2.1
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 1 |
| chr6_+_58569808 | 0.09 |
ENSDART00000154393
|
helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr4_-_57340828 | 0.09 |
ENSDART00000193689
|
zgc:194906
|
zgc:194906 |
| chr22_-_24757785 | 0.08 |
ENSDART00000078225
|
vtg5
|
vitellogenin 5 |
| chr1_+_31054915 | 0.08 |
ENSDART00000148968
|
itga6b
|
integrin, alpha 6b |
| chr2_+_17525355 | 0.08 |
ENSDART00000074101
|
pimr196
|
Pim proto-oncogene, serine/threonine kinase, related 196 |
| chr15_+_41932069 | 0.08 |
ENSDART00000143007
|
si:ch211-191a16.2
|
si:ch211-191a16.2 |
| chr15_-_41762530 | 0.08 |
ENSDART00000187125
ENSDART00000154971 |
ftr91
|
finTRIM family, member 91 |
| chr7_+_20393386 | 0.08 |
ENSDART00000173471
|
si:dkey-33c9.6
|
si:dkey-33c9.6 |
| chr18_+_40643660 | 0.07 |
ENSDART00000134235
|
si:ch211-132b12.6
|
si:ch211-132b12.6 |
| chr16_-_33806390 | 0.07 |
ENSDART00000160671
|
rspo1
|
R-spondin 1 |
| chr12_+_27243059 | 0.07 |
ENSDART00000066269
|
arl4d
|
ADP-ribosylation factor-like 4D |
| chr11_-_5865744 | 0.07 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr4_+_54645654 | 0.06 |
ENSDART00000192864
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr19_+_7453996 | 0.06 |
ENSDART00000147491
ENSDART00000136502 |
mindy1
|
MINDY lysine 48 deubiquitinase 1 |
| chr4_-_42397126 | 0.06 |
ENSDART00000162437
|
si:ch211-129p6.2
|
si:ch211-129p6.2 |
| chr20_+_812012 | 0.06 |
ENSDART00000179299
ENSDART00000137135 |
mei4
|
meiosis-specific, MEI4 homolog (S. cerevisiae) |
| chr15_+_21202820 | 0.06 |
ENSDART00000154036
|
si:dkey-52d15.2
|
si:dkey-52d15.2 |
| chr1_-_30510839 | 0.05 |
ENSDART00000168189
ENSDART00000174868 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr23_-_36449111 | 0.05 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr24_-_37225877 | 0.05 |
ENSDART00000087438
|
cog7
|
component of oligomeric golgi complex 7 |
| chr17_+_25871304 | 0.05 |
ENSDART00000185143
|
wapla
|
WAPL cohesin release factor a |
| chr20_+_12830448 | 0.05 |
ENSDART00000164754
|
BX547930.4
|
|
| chr2_-_38256589 | 0.04 |
ENSDART00000173064
|
si:ch211-14a17.10
|
si:ch211-14a17.10 |
| chr22_-_9973918 | 0.04 |
ENSDART00000146598
ENSDART00000106026 |
nlrc7
|
NLR family CARD domain containing 7 |
| chr16_+_17089114 | 0.04 |
ENSDART00000173223
|
cd27
|
CD27 molecule |
| chr5_-_61797220 | 0.04 |
ENSDART00000079855
|
im:7138535
|
im:7138535 |
| chr11_+_23265157 | 0.04 |
ENSDART00000110152
|
csf1a
|
colony stimulating factor 1a (macrophage) |
| chr2_+_38370535 | 0.04 |
ENSDART00000179614
|
psmb11a
|
proteasome subunit beta 11a |
| chr24_-_42090635 | 0.04 |
ENSDART00000166413
|
ssr1
|
signal sequence receptor, alpha |
| chr5_-_32338866 | 0.03 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr5_+_38685089 | 0.03 |
ENSDART00000139743
|
si:dkey-58f10.10
|
si:dkey-58f10.10 |
| chr6_-_48087152 | 0.03 |
ENSDART00000180614
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr1_-_5745420 | 0.03 |
ENSDART00000166779
|
nrp2a
|
neuropilin 2a |
| chr22_+_28888781 | 0.02 |
ENSDART00000145629
|
pimr98
|
Pim proto-oncogene, serine/threonine kinase, related 98 |
| chr1_-_30457062 | 0.02 |
ENSDART00000185318
ENSDART00000157924 ENSDART00000161380 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr4_-_36032930 | 0.02 |
ENSDART00000191414
|
zgc:174180
|
zgc:174180 |
| chr4_-_70131926 | 0.02 |
ENSDART00000108511
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr24_+_1023839 | 0.01 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr7_-_47282853 | 0.01 |
ENSDART00000169299
|
si:ch211-186j3.3
|
si:ch211-186j3.3 |
| chr22_-_7457247 | 0.01 |
ENSDART00000106081
|
BX511034.2
|
|
| chr7_+_3442834 | 0.01 |
ENSDART00000138247
|
si:ch211-285c6.2
|
si:ch211-285c6.2 |
| chr16_+_23326085 | 0.01 |
ENSDART00000180935
|
krtcap2
|
keratinocyte associated protein 2 |
| chr24_+_12983434 | 0.01 |
ENSDART00000145214
ENSDART00000146911 ENSDART00000066700 |
eloca
|
elongin C paralog a |
| chr4_+_77195833 | 0.01 |
ENSDART00000188365
|
si:dkey-172k15.6
|
si:dkey-172k15.6 |
| chr18_-_6151793 | 0.00 |
ENSDART00000122307
|
gcsha
|
glycine cleavage system protein H (aminomethyl carrier), a |
Network of associatons between targets according to the STRING database.
First level regulatory network of mafa+mafbb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0015840 | urea transport(GO:0015840) |
| 0.4 | 1.7 | GO:0043084 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.3 | 1.1 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 0.6 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 1.5 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 6.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.6 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 1.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.5 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.1 | 11.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 1.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 0.6 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.6 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.1 | 0.4 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.1 | 1.2 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.2 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.1 | 1.8 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 0.1 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 0.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.2 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 1.0 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.4 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 1.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.7 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.6 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 1.6 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.9 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.0 | 0.5 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.7 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.5 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 0.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.7 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.1 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.7 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 1.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 6.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.0 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.7 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.7 | GO:0005770 | late endosome(GO:0005770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.2 | 0.9 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 1.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 11.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.6 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 2.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.5 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.5 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.7 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 1.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.5 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.9 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |