Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
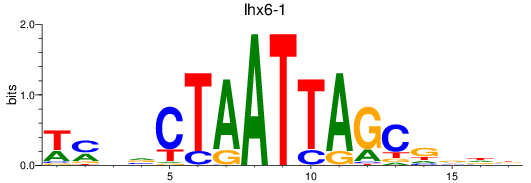
Results for lhx6-1
Z-value: 0.24
Transcription factors associated with lhx6-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx6-1
|
ENSDARG00000052165 | si_ch211-236k19.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch211-236k19.2 | dr11_v1_chr5_-_64900552_64900552 | 0.32 | 1.9e-01 | Click! |
Activity profile of lhx6-1 motif
Sorted Z-values of lhx6-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_20352400 | 0.30 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr4_+_74929427 | 0.23 |
ENSDART00000174082
|
nup50
|
nucleoporin 50 |
| chr20_+_46040666 | 0.21 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr6_+_102506 | 0.17 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr1_-_20928772 | 0.17 |
ENSDART00000078277
|
msmo1
|
methylsterol monooxygenase 1 |
| chr7_+_30392613 | 0.15 |
ENSDART00000075508
|
lipca
|
lipase, hepatic a |
| chr16_-_28658341 | 0.14 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr12_-_32421046 | 0.13 |
ENSDART00000075567
|
enpp7.1
|
ectonucleotide pyrophosphatase/phosphodiesterase 7, tandem duplicate 1 |
| chr16_+_23975930 | 0.12 |
ENSDART00000147858
ENSDART00000144347 ENSDART00000115270 |
apoc4
|
apolipoprotein C-IV |
| chr25_+_3326885 | 0.12 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr9_-_6372535 | 0.12 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr17_-_42988356 | 0.11 |
ENSDART00000024558
|
zgc:92137
|
zgc:92137 |
| chr25_+_3327071 | 0.11 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr14_-_14659023 | 0.11 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr4_+_62431132 | 0.11 |
ENSDART00000160226
|
si:ch211-79g12.1
|
si:ch211-79g12.1 |
| chr25_+_4760489 | 0.10 |
ENSDART00000167399
|
FP245455.1
|
|
| chr13_+_24584401 | 0.10 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr8_+_21353878 | 0.09 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr5_-_23277385 | 0.09 |
ENSDART00000134982
|
plp1b
|
proteolipid protein 1b |
| chr22_-_30770751 | 0.09 |
ENSDART00000172115
|
AL831726.2
|
|
| chr6_-_39344259 | 0.09 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr19_-_617246 | 0.09 |
ENSDART00000062551
|
cyp51
|
cytochrome P450, family 51 |
| chr6_+_3334710 | 0.09 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr19_-_12648122 | 0.08 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr13_+_228045 | 0.08 |
ENSDART00000161091
|
zgc:64201
|
zgc:64201 |
| chr3_-_31079186 | 0.07 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr21_-_39566854 | 0.07 |
ENSDART00000020174
|
dynll2b
|
dynein, light chain, LC8-type 2b |
| chr22_-_5518117 | 0.07 |
ENSDART00000164613
|
CABZ01064972.1
|
|
| chr19_-_12648408 | 0.07 |
ENSDART00000103692
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr12_+_20567382 | 0.07 |
ENSDART00000123737
|
FP885542.1
|
|
| chr3_-_23407720 | 0.07 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr16_+_47207691 | 0.07 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr16_+_23976227 | 0.07 |
ENSDART00000193013
|
apoc4
|
apolipoprotein C-IV |
| chr2_+_10007113 | 0.06 |
ENSDART00000155213
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr19_-_27858033 | 0.06 |
ENSDART00000103898
ENSDART00000144884 |
srd5a1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) |
| chr13_+_9468535 | 0.06 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr13_+_8954667 | 0.06 |
ENSDART00000100241
|
haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr1_+_29068654 | 0.06 |
ENSDART00000053932
|
cbsa
|
cystathionine-beta-synthase a |
| chr2_+_30369116 | 0.06 |
ENSDART00000142137
|
pi15b
|
peptidase inhibitor 15b |
| chr15_+_45563656 | 0.06 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr8_-_45277370 | 0.06 |
ENSDART00000146364
|
adamts13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr11_+_5588122 | 0.06 |
ENSDART00000113281
|
zgc:172302
|
zgc:172302 |
| chr13_-_9045879 | 0.06 |
ENSDART00000155463
ENSDART00000140041 ENSDART00000137454 |
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr16_+_21330634 | 0.06 |
ENSDART00000191285
ENSDART00000183267 |
osbpl3b
|
oxysterol binding protein-like 3b |
| chr2_-_2957970 | 0.06 |
ENSDART00000162505
|
si:ch1073-82l19.1
|
si:ch1073-82l19.1 |
| chr6_-_39764995 | 0.06 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr4_-_71912457 | 0.05 |
ENSDART00000179685
|
si:dkey-92c21.1
|
si:dkey-92c21.1 |
| chr5_+_13394543 | 0.05 |
ENSDART00000051669
ENSDART00000135921 |
tctn2
|
tectonic family member 2 |
| chr7_-_33868903 | 0.05 |
ENSDART00000173500
ENSDART00000178746 |
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr24_+_14581864 | 0.05 |
ENSDART00000134536
|
thtpa
|
thiamine triphosphatase |
| chr12_-_22238004 | 0.05 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr20_+_18661624 | 0.05 |
ENSDART00000152136
ENSDART00000126959 |
tnfaip2a
|
tumor necrosis factor, alpha-induced protein 2a |
| chr6_-_19042294 | 0.05 |
ENSDART00000159461
|
si:rp71-81e14.2
|
si:rp71-81e14.2 |
| chr23_-_37536127 | 0.05 |
ENSDART00000078253
|
tor1l1
|
torsin family 1 like 1 |
| chr17_+_8212477 | 0.05 |
ENSDART00000064665
|
slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr7_+_54642005 | 0.05 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr25_+_34641536 | 0.05 |
ENSDART00000167033
|
CABZ01079011.1
|
|
| chr22_+_19407531 | 0.05 |
ENSDART00000141060
|
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr22_-_15578402 | 0.05 |
ENSDART00000062986
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr2_+_30721466 | 0.05 |
ENSDART00000128982
|
si:dkey-94e7.2
|
si:dkey-94e7.2 |
| chr18_-_48547564 | 0.05 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr24_-_31425799 | 0.05 |
ENSDART00000157998
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr6_-_46861676 | 0.04 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr11_-_44876005 | 0.04 |
ENSDART00000192006
|
opn6a
|
opsin 6, group member a |
| chr16_-_51288178 | 0.04 |
ENSDART00000079864
|
zgc:173729
|
zgc:173729 |
| chr10_-_15963903 | 0.04 |
ENSDART00000142357
|
si:dkey-3h23.3
|
si:dkey-3h23.3 |
| chr1_+_7517454 | 0.04 |
ENSDART00000016139
|
lancl1
|
LanC antibiotic synthetase component C-like 1 (bacterial) |
| chr4_-_32466449 | 0.04 |
ENSDART00000192050
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr12_+_1469327 | 0.04 |
ENSDART00000059143
|
usp22
|
ubiquitin specific peptidase 22 |
| chr18_+_7073130 | 0.04 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr5_+_19479200 | 0.04 |
ENSDART00000137703
|
aldh3b2
|
aldehyde dehydrogenase 3 family, member B2 |
| chr8_-_23780334 | 0.04 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr20_-_53366137 | 0.04 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr13_-_9213207 | 0.04 |
ENSDART00000139861
ENSDART00000140524 |
HTRA2 (1 of many)
|
si:dkey-33c12.11 |
| chr5_-_23843636 | 0.04 |
ENSDART00000193280
|
GBGT1 (1 of many)
|
si:ch211-135f11.5 |
| chr2_+_56937548 | 0.04 |
ENSDART00000189308
|
CABZ01117752.1
|
|
| chr19_+_1673599 | 0.04 |
ENSDART00000163127
|
klhl7
|
kelch-like family member 7 |
| chr1_+_19708508 | 0.04 |
ENSDART00000054581
ENSDART00000131206 |
march1
|
membrane-associated ring finger (C3HC4) 1 |
| chr13_+_9496748 | 0.04 |
ENSDART00000147050
ENSDART00000102120 |
CR848040.4
|
|
| chr24_-_26386627 | 0.04 |
ENSDART00000125344
|
skilb
|
SKI-like proto-oncogene b |
| chr4_+_2637947 | 0.04 |
ENSDART00000130623
|
dus4l
|
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr4_-_2440175 | 0.04 |
ENSDART00000058059
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr22_+_19366866 | 0.04 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr3_-_32934359 | 0.04 |
ENSDART00000124160
|
aoc2
|
amine oxidase, copper containing 2 |
| chr16_+_39159752 | 0.04 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr24_-_32582378 | 0.04 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr21_+_28958471 | 0.04 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr14_+_7902374 | 0.04 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr6_+_120181 | 0.04 |
ENSDART00000151209
ENSDART00000185930 |
cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr5_-_7199998 | 0.04 |
ENSDART00000167316
|
CABZ01087502.1
|
|
| chr4_-_28158335 | 0.04 |
ENSDART00000134605
|
gramd4a
|
GRAM domain containing 4a |
| chr19_+_9295244 | 0.04 |
ENSDART00000132255
ENSDART00000144299 |
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr8_-_45867358 | 0.03 |
ENSDART00000132810
|
adam9
|
ADAM metallopeptidase domain 9 |
| chr5_+_7564644 | 0.03 |
ENSDART00000192173
|
CABZ01039096.1
|
|
| chr2_-_5561005 | 0.03 |
ENSDART00000057439
ENSDART00000137882 |
parla
|
presenilin associated, rhomboid-like a |
| chr25_-_10791437 | 0.03 |
ENSDART00000127054
|
BX572619.1
|
|
| chr5_-_66301142 | 0.03 |
ENSDART00000067541
|
prlrb
|
prolactin receptor b |
| chr21_-_5205617 | 0.03 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr13_+_49175947 | 0.03 |
ENSDART00000056927
|
egln1a
|
egl-9 family hypoxia-inducible factor 1a |
| chr4_-_837768 | 0.03 |
ENSDART00000185280
ENSDART00000135618 |
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr1_+_55755304 | 0.03 |
ENSDART00000144983
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr4_+_9467049 | 0.03 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr16_-_32013913 | 0.03 |
ENSDART00000030282
ENSDART00000138701 |
gstk1
|
glutathione S-transferase kappa 1 |
| chr16_+_42471455 | 0.03 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr17_+_30704068 | 0.03 |
ENSDART00000062793
|
apoba
|
apolipoprotein Ba |
| chr7_+_20344032 | 0.03 |
ENSDART00000144948
ENSDART00000138786 |
ponzr1
|
plac8 onzin related protein 1 |
| chr15_+_37954666 | 0.03 |
ENSDART00000126534
ENSDART00000155548 |
si:dkey-238d18.7
|
si:dkey-238d18.7 |
| chr15_-_20956384 | 0.03 |
ENSDART00000135770
|
tbcela
|
tubulin folding cofactor E-like a |
| chr19_+_2631565 | 0.03 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr18_+_1145571 | 0.03 |
ENSDART00000135055
|
rec114
|
REC114 meiotic recombination protein |
| chr8_+_25767610 | 0.03 |
ENSDART00000062406
|
cacna1sb
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, b |
| chr18_-_14274803 | 0.03 |
ENSDART00000166643
|
mlycd
|
malonyl-CoA decarboxylase |
| chr19_+_42983613 | 0.03 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr2_-_15031858 | 0.03 |
ENSDART00000191478
|
hccsa.1
|
holocytochrome c synthase a |
| chr9_-_23994225 | 0.03 |
ENSDART00000140346
|
col6a3
|
collagen, type VI, alpha 3 |
| chr13_+_29470442 | 0.03 |
ENSDART00000028417
|
lrit2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr7_+_20344222 | 0.03 |
ENSDART00000141186
ENSDART00000139274 |
ponzr1
|
plac8 onzin related protein 1 |
| chr4_+_77957611 | 0.03 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr13_+_49175624 | 0.03 |
ENSDART00000193550
|
egln1a
|
egl-9 family hypoxia-inducible factor 1a |
| chr5_-_40292299 | 0.03 |
ENSDART00000097525
|
snx18a
|
sorting nexin 18a |
| chr6_+_13779857 | 0.03 |
ENSDART00000154793
|
tmem198b
|
transmembrane protein 198b |
| chr10_+_44584614 | 0.03 |
ENSDART00000163523
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr1_-_1402303 | 0.03 |
ENSDART00000130697
|
runx1
|
runt-related transcription factor 1 |
| chr2_-_55298075 | 0.03 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr2_+_30721070 | 0.03 |
ENSDART00000099052
|
si:dkey-94e7.2
|
si:dkey-94e7.2 |
| chr2_+_30368800 | 0.03 |
ENSDART00000179564
|
pi15b
|
peptidase inhibitor 15b |
| chr20_-_47188966 | 0.03 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr7_+_50822465 | 0.02 |
ENSDART00000011082
|
per1b
|
period circadian clock 1b |
| chr4_+_29967255 | 0.02 |
ENSDART00000150383
|
znf1128
|
zinc finger protein 1128 |
| chr10_-_3138403 | 0.02 |
ENSDART00000183365
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr15_+_33691455 | 0.02 |
ENSDART00000170329
|
hdac12
|
histone deacetylase 12 |
| chr2_-_27329667 | 0.02 |
ENSDART00000187490
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr17_-_6198823 | 0.02 |
ENSDART00000028407
ENSDART00000193636 |
ptk2ba
|
protein tyrosine kinase 2 beta, a |
| chr22_-_21897203 | 0.02 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr12_-_18883079 | 0.02 |
ENSDART00000178619
|
shisa8b
|
shisa family member 8b |
| chr18_-_25568994 | 0.02 |
ENSDART00000133029
|
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr12_-_36521767 | 0.02 |
ENSDART00000110290
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr10_+_34175476 | 0.02 |
ENSDART00000102123
|
zgc:174193
|
zgc:174193 |
| chr4_-_71152937 | 0.02 |
ENSDART00000191017
|
si:dkey-193i10.4
|
si:dkey-193i10.4 |
| chr13_+_9521629 | 0.02 |
ENSDART00000149870
ENSDART00000137666 |
si:dkey-19p15.4
|
si:dkey-19p15.4 |
| chr20_+_27691307 | 0.02 |
ENSDART00000153299
|
si:dkey-1h6.8
|
si:dkey-1h6.8 |
| chr12_-_33357655 | 0.02 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr17_-_37195163 | 0.02 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr3_-_10751491 | 0.02 |
ENSDART00000016351
|
zgc:112965
|
zgc:112965 |
| chr19_-_31341850 | 0.02 |
ENSDART00000040810
|
arl4ab
|
ADP-ribosylation factor-like 4ab |
| chr24_+_38534550 | 0.02 |
ENSDART00000105677
|
zgc:154125
|
zgc:154125 |
| chr20_+_19006703 | 0.02 |
ENSDART00000128435
|
pinx1
|
PIN2/TERF1 interacting, telomerase inhibitor 1 |
| chr2_-_21621878 | 0.02 |
ENSDART00000189540
|
zgc:55781
|
zgc:55781 |
| chr4_-_65525733 | 0.02 |
ENSDART00000159401
|
BX293993.1
|
|
| chr7_+_69459759 | 0.02 |
ENSDART00000160500
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr4_+_62576422 | 0.02 |
ENSDART00000180679
|
si:ch211-79g12.2
|
si:ch211-79g12.2 |
| chr16_-_11798994 | 0.02 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr3_-_13147310 | 0.02 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr10_-_3138859 | 0.02 |
ENSDART00000190606
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr24_-_24724233 | 0.02 |
ENSDART00000127044
ENSDART00000012399 |
armc1
|
armadillo repeat containing 1 |
| chr2_+_21312972 | 0.02 |
ENSDART00000080495
|
zbtb47a
|
zinc finger and BTB domain containing 47a |
| chr1_-_44161417 | 0.02 |
ENSDART00000083213
|
slc43a3a
|
solute carrier family 43, member 3a |
| chr8_-_979735 | 0.02 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr17_+_38030327 | 0.02 |
ENSDART00000085481
|
slc25a21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr19_-_6134802 | 0.02 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr15_-_2497568 | 0.02 |
ENSDART00000080398
|
neu4
|
sialidase 4 |
| chr22_-_27709692 | 0.02 |
ENSDART00000172458
|
CR547131.1
|
|
| chr9_-_12886108 | 0.02 |
ENSDART00000177283
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr22_-_17729778 | 0.02 |
ENSDART00000192132
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr11_+_4026229 | 0.02 |
ENSDART00000041417
|
camk1b
|
calcium/calmodulin-dependent protein kinase Ib |
| chr6_-_54815886 | 0.02 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr22_+_6711549 | 0.02 |
ENSDART00000132094
|
CT583625.4
|
|
| chr19_-_1871415 | 0.02 |
ENSDART00000004585
|
clptm1l
|
CLPTM1-like |
| chr22_+_6587280 | 0.02 |
ENSDART00000106185
|
CR352226.7
|
|
| chr7_-_17690983 | 0.02 |
ENSDART00000009367
|
map4k2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr23_-_41651759 | 0.02 |
ENSDART00000146808
|
si:ch73-184c24.1
|
si:ch73-184c24.1 |
| chr21_+_4328348 | 0.02 |
ENSDART00000131658
ENSDART00000143799 |
HTRA2 (1 of many)
|
si:dkey-84o3.7 |
| chr20_-_52642504 | 0.02 |
ENSDART00000138982
ENSDART00000134999 |
si:ch211-221n20.2
|
si:ch211-221n20.2 |
| chr4_+_11384891 | 0.02 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr17_-_37195354 | 0.02 |
ENSDART00000190963
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr4_-_29753895 | 0.02 |
ENSDART00000150742
|
si:dkey-191j3.1
|
si:dkey-191j3.1 |
| chr13_+_771403 | 0.02 |
ENSDART00000093166
|
nrxn1b
|
neurexin 1b |
| chr2_-_30721502 | 0.02 |
ENSDART00000132389
|
si:dkey-94e7.1
|
si:dkey-94e7.1 |
| chr15_-_18200358 | 0.02 |
ENSDART00000158569
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr22_+_6727266 | 0.02 |
ENSDART00000142352
|
CT583625.6
|
|
| chr25_+_21324588 | 0.02 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr2_-_3038904 | 0.02 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
| chr4_+_41789497 | 0.02 |
ENSDART00000126634
|
si:dkey-237m9.2
|
si:dkey-237m9.2 |
| chr6_+_9870192 | 0.02 |
ENSDART00000150894
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr21_+_6751760 | 0.02 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr2_+_24936766 | 0.02 |
ENSDART00000025962
|
gyg1a
|
glycogenin 1a |
| chr16_-_51888952 | 0.02 |
ENSDART00000186407
ENSDART00000175435 |
rpl28
|
ribosomal protein L28 |
| chr19_-_46088429 | 0.02 |
ENSDART00000161385
|
ptdss1b
|
phosphatidylserine synthase 1b |
| chr12_-_36521947 | 0.02 |
ENSDART00000152946
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr6_+_34511886 | 0.02 |
ENSDART00000179450
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr4_+_11375894 | 0.02 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr11_+_29790626 | 0.02 |
ENSDART00000067822
|
dynlt3
|
dynein, light chain, Tctex-type 3 |
| chr19_-_22541284 | 0.02 |
ENSDART00000019505
|
pleca
|
plectin a |
| chr1_-_29139141 | 0.02 |
ENSDART00000075546
ENSDART00000133246 |
hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr25_+_13498188 | 0.02 |
ENSDART00000015710
|
snrkb
|
SNF related kinase b |
| chr19_-_4793263 | 0.02 |
ENSDART00000147510
ENSDART00000141336 ENSDART00000110551 ENSDART00000146684 |
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr11_+_7264457 | 0.01 |
ENSDART00000154182
|
reep6
|
receptor accessory protein 6 |
| chr22_-_10121880 | 0.01 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr3_-_23406964 | 0.01 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr5_-_9625459 | 0.01 |
ENSDART00000143347
|
sh2b3
|
SH2B adaptor protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx6-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.1 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.0 | 0.1 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.1 | GO:0035671 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.1 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |