Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
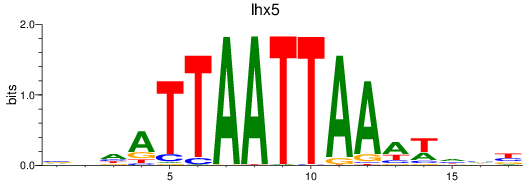
Results for lhx5
Z-value: 0.68
Transcription factors associated with lhx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx5
|
ENSDARG00000057936 | LIM homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx5 | dr11_v1_chr21_-_15929041_15929041 | -0.39 | 1.0e-01 | Click! |
Activity profile of lhx5 motif
Sorted Z-values of lhx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22099536 | 2.55 |
ENSDART00000101923
|
CR391987.1
|
|
| chr6_+_7444899 | 2.46 |
ENSDART00000053775
|
arf3b
|
ADP-ribosylation factor 3b |
| chr17_-_37395460 | 2.28 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr3_-_11624694 | 2.12 |
ENSDART00000127157
|
hlfa
|
hepatic leukemia factor a |
| chr25_+_31227747 | 1.87 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr5_+_2815021 | 1.86 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr16_-_28658341 | 1.84 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr3_-_59297532 | 1.75 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr19_-_26869103 | 1.69 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr15_-_37850969 | 1.64 |
ENSDART00000031418
|
hsc70
|
heat shock cognate 70 |
| chr15_-_44601331 | 1.64 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr3_-_48716422 | 1.58 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr19_+_43297546 | 1.55 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr16_+_2820340 | 1.52 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr9_-_22147567 | 1.45 |
ENSDART00000110941
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr21_+_21195487 | 1.44 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr15_-_46718759 | 1.43 |
ENSDART00000085926
ENSDART00000154577 |
zgc:162872
|
zgc:162872 |
| chr6_+_39905021 | 1.41 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr20_-_2949028 | 1.36 |
ENSDART00000104667
ENSDART00000193151 ENSDART00000131946 |
cdk19
|
cyclin-dependent kinase 19 |
| chr10_+_45089820 | 1.35 |
ENSDART00000175481
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr9_+_34641237 | 1.34 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr25_-_19420949 | 1.29 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr12_-_17712393 | 1.25 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr10_-_2524297 | 1.15 |
ENSDART00000192475
|
CU856539.1
|
|
| chr9_+_2574122 | 1.13 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr17_-_200316 | 1.10 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr19_+_5480327 | 1.09 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr24_-_11309477 | 1.09 |
ENSDART00000137257
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr7_-_71829649 | 1.08 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr4_-_18309917 | 1.06 |
ENSDART00000189084
|
plxnc1
|
plexin C1 |
| chr16_-_38118003 | 1.06 |
ENSDART00000058667
|
si:dkey-23o4.6
|
si:dkey-23o4.6 |
| chr13_-_29421331 | 1.05 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr23_-_17509656 | 1.05 |
ENSDART00000148423
|
dnajc5ab
|
DnaJ (Hsp40) homolog, subfamily C, member 5ab |
| chr10_+_17776981 | 1.04 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr10_+_38417512 | 1.02 |
ENSDART00000112457
|
samsn1b
|
SAM domain, SH3 domain and nuclear localisation signals 1b |
| chr21_-_20939488 | 1.00 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr23_-_18024543 | 1.00 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr20_+_52389858 | 0.99 |
ENSDART00000185863
ENSDART00000166651 |
arhgap39
|
Rho GTPase activating protein 39 |
| chr9_+_54039006 | 0.99 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr18_+_15644559 | 0.98 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr7_-_71531846 | 0.98 |
ENSDART00000111797
|
acer2
|
alkaline ceramidase 2 |
| chr5_-_6377865 | 0.97 |
ENSDART00000031775
|
zgc:73226
|
zgc:73226 |
| chr12_-_6880694 | 0.96 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr6_-_28980756 | 0.95 |
ENSDART00000014661
|
glmnb
|
glomulin, FKBP associated protein b |
| chr7_+_24573721 | 0.94 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr20_+_32552912 | 0.93 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr23_+_16638639 | 0.92 |
ENSDART00000143545
|
snphb
|
syntaphilin b |
| chr16_-_28856112 | 0.92 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr5_-_41494831 | 0.91 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr24_-_7697274 | 0.90 |
ENSDART00000186077
|
syt5b
|
synaptotagmin Vb |
| chr16_+_51326237 | 0.88 |
ENSDART00000168525
|
CU469384.1
|
|
| chr8_+_52637507 | 0.86 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr17_-_23616626 | 0.84 |
ENSDART00000104730
|
ifit14
|
interferon-induced protein with tetratricopeptide repeats 14 |
| chr19_-_47323267 | 0.82 |
ENSDART00000190077
|
CU138512.1
|
|
| chr12_+_45200744 | 0.81 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr23_-_1660708 | 0.79 |
ENSDART00000175138
|
CU693481.1
|
|
| chr4_-_2219705 | 0.79 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr6_-_43677125 | 0.79 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr17_+_16564921 | 0.78 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr5_-_21422390 | 0.77 |
ENSDART00000144198
|
tenm1
|
teneurin transmembrane protein 1 |
| chr1_+_6862917 | 0.76 |
ENSDART00000182953
|
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr6_-_57539141 | 0.76 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr25_+_3994823 | 0.75 |
ENSDART00000154020
|
eps8l2
|
EPS8 like 2 |
| chr17_-_16965809 | 0.75 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr2_+_2223837 | 0.74 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr9_+_54290896 | 0.73 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr2_-_9059955 | 0.73 |
ENSDART00000022768
|
ak5
|
adenylate kinase 5 |
| chr23_-_39666519 | 0.73 |
ENSDART00000110868
ENSDART00000190961 |
vwa1
|
von Willebrand factor A domain containing 1 |
| chr3_+_39853788 | 0.73 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr7_-_74057288 | 0.72 |
ENSDART00000189740
|
CU929544.1
|
|
| chr11_-_45141309 | 0.72 |
ENSDART00000181736
|
cant1b
|
calcium activated nucleotidase 1b |
| chr7_-_66868543 | 0.71 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr20_-_5291012 | 0.70 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr15_-_46779934 | 0.70 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr2_+_20430366 | 0.70 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr21_-_44081540 | 0.69 |
ENSDART00000130833
|
FO704810.1
|
|
| chr9_+_38074082 | 0.68 |
ENSDART00000017833
|
cacnb4a
|
calcium channel, voltage-dependent, beta 4a subunit |
| chr17_+_28533102 | 0.65 |
ENSDART00000156218
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr6_-_1865323 | 0.65 |
ENSDART00000155775
|
dlgap4b
|
discs, large (Drosophila) homolog-associated protein 4b |
| chr23_+_26946429 | 0.65 |
ENSDART00000185564
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr24_-_31425799 | 0.65 |
ENSDART00000157998
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr22_-_14739491 | 0.65 |
ENSDART00000133385
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr9_+_7998794 | 0.64 |
ENSDART00000138167
|
myo16
|
myosin XVI |
| chr15_+_9327252 | 0.64 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr1_-_55785722 | 0.63 |
ENSDART00000142069
ENSDART00000043933 |
ndufb7
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 |
| chr22_-_31060579 | 0.63 |
ENSDART00000182376
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr25_-_27621268 | 0.63 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr6_-_39649504 | 0.63 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr4_-_75678082 | 0.61 |
ENSDART00000180201
|
si:dkey-165o8.1
|
si:dkey-165o8.1 |
| chr2_+_42072231 | 0.61 |
ENSDART00000084517
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr25_-_25058508 | 0.61 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr10_+_16584382 | 0.60 |
ENSDART00000112039
|
CR790388.1
|
|
| chr1_+_54124209 | 0.60 |
ENSDART00000187730
|
LO017722.1
|
|
| chr24_+_9475809 | 0.60 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr23_+_31405497 | 0.59 |
ENSDART00000053546
|
sh3bgrl2
|
SH3 domain binding glutamate-rich protein like 2 |
| chr12_+_18681477 | 0.59 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr1_-_15797663 | 0.59 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr9_+_24159280 | 0.58 |
ENSDART00000184624
ENSDART00000178422 |
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr17_+_38476300 | 0.58 |
ENSDART00000123298
|
stard9
|
StAR-related lipid transfer (START) domain containing 9 |
| chr5_+_69747417 | 0.58 |
ENSDART00000153717
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr16_-_31622777 | 0.57 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr15_+_34592215 | 0.57 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr1_+_44127292 | 0.56 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr3_+_45687266 | 0.56 |
ENSDART00000131652
|
gpr146
|
G protein-coupled receptor 146 |
| chr15_-_17618800 | 0.55 |
ENSDART00000157185
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr15_-_40267485 | 0.54 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr25_-_9805269 | 0.54 |
ENSDART00000192048
|
lrrc4c
|
leucine rich repeat containing 4C |
| chr21_-_22635245 | 0.54 |
ENSDART00000115224
ENSDART00000101782 |
nectin1a
|
nectin cell adhesion molecule 1a |
| chr4_+_20536120 | 0.52 |
ENSDART00000125685
|
zmp:0000000650
|
zmp:0000000650 |
| chr3_+_7808459 | 0.51 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr1_-_50438247 | 0.51 |
ENSDART00000114098
|
dkk2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr19_-_5769728 | 0.51 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr21_+_27278120 | 0.51 |
ENSDART00000193882
|
si:dkey-175m17.7
|
si:dkey-175m17.7 |
| chr11_-_26832685 | 0.50 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr10_-_2524917 | 0.49 |
ENSDART00000188642
|
CU856539.1
|
|
| chr20_-_43663494 | 0.48 |
ENSDART00000144564
|
BX470188.1
|
|
| chr4_-_5019113 | 0.48 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr22_+_30047245 | 0.48 |
ENSDART00000142857
ENSDART00000141247 ENSDART00000140015 ENSDART00000040538 |
add3a
|
adducin 3 (gamma) a |
| chr9_+_24159725 | 0.46 |
ENSDART00000137756
|
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr10_-_43404027 | 0.46 |
ENSDART00000086227
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr19_-_5769553 | 0.46 |
ENSDART00000175003
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr7_+_59212666 | 0.46 |
ENSDART00000172046
|
dok1b
|
docking protein 1b |
| chr21_-_39327223 | 0.45 |
ENSDART00000115097
|
aifm5
|
apoptosis-inducing factor, mitochondrion-associated, 5 |
| chr17_-_40956035 | 0.44 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr12_+_1139690 | 0.44 |
ENSDART00000160442
|
CABZ01072885.1
|
|
| chr24_-_33873451 | 0.44 |
ENSDART00000159840
|
asic1c
|
acid-sensing (proton-gated) ion channel 1c |
| chr5_-_48664522 | 0.44 |
ENSDART00000083229
|
mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr12_-_48006835 | 0.44 |
ENSDART00000108989
|
adamts14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14 |
| chr5_-_38094130 | 0.43 |
ENSDART00000131831
|
si:ch211-284e13.4
|
si:ch211-284e13.4 |
| chr16_-_41004731 | 0.42 |
ENSDART00000102591
|
KCNV1
|
si:dkey-201i6.2 |
| chr8_+_36500061 | 0.42 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr10_-_13343831 | 0.41 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr18_+_35128685 | 0.41 |
ENSDART00000151579
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr5_+_13427826 | 0.41 |
ENSDART00000083359
|
sec14l8
|
SEC14-like lipid binding 8 |
| chr22_+_19366866 | 0.41 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr9_-_9415000 | 0.40 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr20_-_13774826 | 0.39 |
ENSDART00000063831
|
opn8c
|
opsin 8, group member c |
| chr15_-_14552101 | 0.39 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr14_-_32631013 | 0.39 |
ENSDART00000176815
|
atp11c
|
ATPase phospholipid transporting 11C |
| chr14_-_48765262 | 0.39 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr17_+_49281597 | 0.38 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr16_-_25829779 | 0.38 |
ENSDART00000086301
|
irge4
|
immunity-related GTPase family, e4 |
| chr11_-_45140227 | 0.38 |
ENSDART00000184062
|
cant1b
|
calcium activated nucleotidase 1b |
| chr8_+_7801060 | 0.37 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr14_-_32631519 | 0.37 |
ENSDART00000167282
ENSDART00000052938 |
atp11c
|
ATPase phospholipid transporting 11C |
| chr21_-_2814709 | 0.36 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr11_+_40812590 | 0.36 |
ENSDART00000186690
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr6_+_52350443 | 0.36 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr24_-_35767501 | 0.36 |
ENSDART00000105680
ENSDART00000042290 ENSDART00000166264 |
dtna
|
dystrobrevin, alpha |
| chr8_-_25569920 | 0.36 |
ENSDART00000136869
|
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr13_-_9886579 | 0.35 |
ENSDART00000101926
|
si:ch211-117n7.7
|
si:ch211-117n7.7 |
| chr1_-_9195629 | 0.35 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr14_+_36521005 | 0.34 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr23_+_39970425 | 0.34 |
ENSDART00000165376
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr2_+_49799470 | 0.34 |
ENSDART00000146325
|
si:ch211-190k17.19
|
si:ch211-190k17.19 |
| chr9_-_37749973 | 0.33 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr8_+_36560019 | 0.33 |
ENSDART00000136418
ENSDART00000061378 ENSDART00000185237 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr23_-_20051369 | 0.33 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr17_-_45040813 | 0.33 |
ENSDART00000075514
|
entpd5a
|
ectonucleoside triphosphate diphosphohydrolase 5a |
| chr9_-_42418470 | 0.32 |
ENSDART00000144353
|
calcrla
|
calcitonin receptor-like a |
| chr23_+_42810055 | 0.32 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr22_-_13466246 | 0.32 |
ENSDART00000134035
|
cntnap5b
|
contactin associated protein-like 5b |
| chr5_-_42904329 | 0.32 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr11_+_30057762 | 0.31 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr25_+_4751879 | 0.31 |
ENSDART00000169465
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr11_-_34478225 | 0.29 |
ENSDART00000189604
|
xxylt1
|
xyloside xylosyltransferase 1 |
| chr14_-_7207961 | 0.29 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr15_-_22074315 | 0.29 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr1_+_51721851 | 0.28 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr11_+_45436703 | 0.28 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr7_-_7692992 | 0.27 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr12_+_7497882 | 0.26 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr4_-_1801519 | 0.25 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr21_-_35534401 | 0.25 |
ENSDART00000112308
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr10_-_11261565 | 0.24 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr8_-_31062811 | 0.24 |
ENSDART00000142528
|
slc20a1a
|
solute carrier family 20, member 1a |
| chr9_-_50001606 | 0.23 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr14_-_451555 | 0.23 |
ENSDART00000190906
|
FAT4
|
FAT atypical cadherin 4 |
| chr22_-_20812822 | 0.22 |
ENSDART00000193778
|
dot1l
|
DOT1-like histone H3K79 methyltransferase |
| chr9_+_55154414 | 0.22 |
ENSDART00000182924
|
ANOS1
|
anosmin 1 |
| chr8_+_36500308 | 0.20 |
ENSDART00000098701
|
slc7a4
|
solute carrier family 7, member 4 |
| chr1_+_41498188 | 0.20 |
ENSDART00000191934
ENSDART00000146310 |
dtx4
|
deltex 4, E3 ubiquitin ligase |
| chr8_-_11834599 | 0.20 |
ENSDART00000190986
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr22_+_19552987 | 0.19 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr13_-_2010191 | 0.19 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr4_+_12966640 | 0.19 |
ENSDART00000113357
|
vhll
|
von Hippel-Lindau tumor suppressor like |
| chr23_+_41800052 | 0.19 |
ENSDART00000141484
|
pdyn
|
prodynorphin |
| chr9_-_12811936 | 0.19 |
ENSDART00000188490
|
myo10l3
|
myosin X-like 3 |
| chr4_+_2482046 | 0.18 |
ENSDART00000103371
|
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr8_+_43340995 | 0.17 |
ENSDART00000038566
|
rflna
|
refilin A |
| chr2_+_20605925 | 0.16 |
ENSDART00000191510
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr11_-_44979281 | 0.16 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr7_-_7692723 | 0.16 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr18_+_5172848 | 0.15 |
ENSDART00000190642
|
CABZ01080599.1
|
|
| chr4_+_5255041 | 0.15 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr14_+_16151368 | 0.15 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr17_-_27048537 | 0.15 |
ENSDART00000050018
ENSDART00000193861 |
cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr11_-_2838699 | 0.15 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr4_+_9669717 | 0.15 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr7_-_50410524 | 0.15 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.4 | 1.6 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.4 | 1.1 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.3 | 1.0 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.3 | 1.9 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.3 | 0.8 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.2 | 1.0 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.2 | 1.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 0.7 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.2 | 0.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 0.9 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.2 | 0.5 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.2 | 0.6 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 0.4 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 0.5 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 1.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.0 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 1.6 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 1.0 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 0.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.0 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.3 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.3 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 2.0 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 0.7 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.0 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 1.8 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.2 | GO:1903011 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.1 | 0.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.1 | 0.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 1.1 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.3 | GO:0071543 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 2.0 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.8 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.7 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.7 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 1.3 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 2.4 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.3 | GO:0055062 | phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.0 | 0.8 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 1.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.6 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 1.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 1.2 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.8 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.4 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.0 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 0.9 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.1 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.6 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.9 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 3.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.6 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 1.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.9 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 2.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.0 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.4 | GO:0030424 | axon(GO:0030424) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.6 | 1.8 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.3 | 1.0 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.3 | 0.8 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 1.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.7 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.1 | 1.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.3 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.1 | 0.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 1.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.3 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 0.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.4 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 1.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.6 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.2 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 1.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.3 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.6 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 1.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 1.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 1.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.7 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 1.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.2 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.9 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 2.1 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 0.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 3.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |