Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
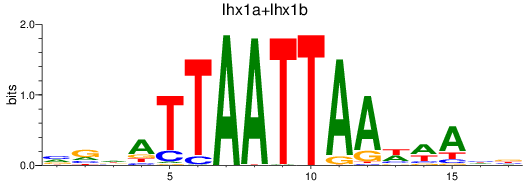
Results for lhx1a+lhx1b
Z-value: 0.25
Transcription factors associated with lhx1a+lhx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx1b
|
ENSDARG00000007944 | LIM homeobox 1b |
|
lhx1a
|
ENSDARG00000014018 | LIM homeobox 1a |
|
lhx1b
|
ENSDARG00000111635 | LIM homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx1b | dr11_v1_chr5_+_56268436_56268436 | 0.58 | 9.9e-03 | Click! |
| lhx1a | dr11_v1_chr15_-_27710513_27710575 | -0.19 | 4.5e-01 | Click! |
Activity profile of lhx1a+lhx1b motif
Sorted Z-values of lhx1a+lhx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_16422654 | 0.96 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr7_-_17297156 | 0.63 |
ENSDART00000161336
|
nitr11a
|
novel immune-type receptor 11a |
| chr3_-_19368435 | 0.48 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr4_+_57881965 | 0.35 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr16_+_54209504 | 0.34 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr6_-_50730749 | 0.34 |
ENSDART00000157153
ENSDART00000110441 |
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr3_+_40164129 | 0.33 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr9_-_27398369 | 0.33 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr2_+_38373272 | 0.31 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr7_+_20471315 | 0.31 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr3_-_15999501 | 0.30 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr24_-_38657683 | 0.30 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr3_-_26806032 | 0.29 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr22_-_14247276 | 0.28 |
ENSDART00000033332
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr5_+_60590796 | 0.28 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr3_-_26787430 | 0.28 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr25_-_21031007 | 0.28 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr23_+_20640484 | 0.28 |
ENSDART00000054691
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr1_-_669717 | 0.27 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr20_-_45812144 | 0.27 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr12_+_13118540 | 0.27 |
ENSDART00000077840
ENSDART00000127870 |
cmn
|
calymmin |
| chr11_-_10456553 | 0.26 |
ENSDART00000169509
ENSDART00000185574 ENSDART00000188276 |
ect2
|
epithelial cell transforming 2 |
| chr5_+_41477954 | 0.26 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr24_+_21514283 | 0.26 |
ENSDART00000007066
|
cdk8
|
cyclin-dependent kinase 8 |
| chr2_-_30668580 | 0.26 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr1_-_45616470 | 0.25 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr19_+_7424347 | 0.25 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr4_+_77943184 | 0.25 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr22_-_5252005 | 0.25 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr9_+_37152564 | 0.24 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
| chr2_+_105748 | 0.24 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr12_-_35830625 | 0.24 |
ENSDART00000180028
|
CU459056.1
|
|
| chr14_+_16287968 | 0.24 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr11_-_10456387 | 0.24 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr23_-_16485190 | 0.24 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr6_-_12912606 | 0.23 |
ENSDART00000164640
|
ical1
|
islet cell autoantigen 1-like |
| chr6_-_34838397 | 0.23 |
ENSDART00000060169
ENSDART00000169605 |
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr5_+_41477526 | 0.22 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr2_+_25315591 | 0.22 |
ENSDART00000161386
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr25_+_22320738 | 0.21 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr13_+_11440389 | 0.21 |
ENSDART00000186463
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr21_+_42717424 | 0.21 |
ENSDART00000166936
ENSDART00000172135 |
sh3pxd2b
|
SH3 and PX domains 2B |
| chr10_+_17235370 | 0.21 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr1_+_21731382 | 0.20 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr21_+_25802190 | 0.19 |
ENSDART00000128987
|
nf2b
|
neurofibromin 2b (merlin) |
| chr15_+_9861973 | 0.19 |
ENSDART00000170945
|
si:dkey-13m3.2
|
si:dkey-13m3.2 |
| chr20_+_11731039 | 0.19 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr3_+_26342768 | 0.19 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr18_+_46382484 | 0.19 |
ENSDART00000024202
ENSDART00000142790 |
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr9_-_14504834 | 0.19 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr20_-_9095105 | 0.19 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr22_-_24285432 | 0.18 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr20_+_25586099 | 0.18 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr8_+_50953776 | 0.18 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr22_-_22242884 | 0.17 |
ENSDART00000020937
|
hdgfl2
|
HDGF like 2 |
| chr6_-_35052145 | 0.17 |
ENSDART00000073970
ENSDART00000185790 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr7_+_22809905 | 0.17 |
ENSDART00000166900
ENSDART00000143455 ENSDART00000126037 |
sf1
|
splicing factor 1 |
| chr5_-_31901468 | 0.17 |
ENSDART00000147814
ENSDART00000141446 |
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr17_+_24318753 | 0.17 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr18_+_14619544 | 0.17 |
ENSDART00000010821
|
utp4
|
UTP4, small subunit processome component |
| chr6_+_11989537 | 0.17 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr16_-_42965192 | 0.16 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr1_-_21287724 | 0.16 |
ENSDART00000193900
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr24_+_21540842 | 0.15 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr1_+_10318089 | 0.15 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr15_-_35112937 | 0.15 |
ENSDART00000154565
ENSDART00000099642 |
zgc:77118
|
zgc:77118 |
| chr16_-_16761164 | 0.15 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr21_-_17482465 | 0.15 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr4_-_9891874 | 0.14 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr23_+_12406722 | 0.14 |
ENSDART00000145008
|
pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr8_-_53926228 | 0.14 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr6_-_55399214 | 0.13 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr18_+_8320165 | 0.13 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr4_+_69191065 | 0.13 |
ENSDART00000170595
|
znf1075
|
zinc finger protein 1075 |
| chr6_-_44402358 | 0.13 |
ENSDART00000193007
ENSDART00000193603 |
pdzrn3b
|
PDZ domain containing RING finger 3b |
| chr21_-_25801956 | 0.13 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr2_+_10878406 | 0.12 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr1_-_51038885 | 0.12 |
ENSDART00000035150
|
spast
|
spastin |
| chr14_+_44794936 | 0.12 |
ENSDART00000128881
|
zgc:195212
|
zgc:195212 |
| chr4_+_38188797 | 0.12 |
ENSDART00000133264
|
si:ch211-229l10.11
|
si:ch211-229l10.11 |
| chr23_-_16484383 | 0.12 |
ENSDART00000187839
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr22_-_36530902 | 0.12 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr8_-_16712111 | 0.11 |
ENSDART00000184147
ENSDART00000180419 ENSDART00000076600 |
rpe65c
|
retinal pigment epithelium-specific protein 65c |
| chr8_-_49728590 | 0.11 |
ENSDART00000135714
ENSDART00000138810 ENSDART00000098319 |
gkap1
|
G kinase anchoring protein 1 |
| chr20_-_28642061 | 0.11 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr9_-_43644261 | 0.11 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr11_-_41853874 | 0.11 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr22_-_10156581 | 0.11 |
ENSDART00000168304
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr1_+_6172786 | 0.11 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr19_+_2631565 | 0.10 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr21_+_6151953 | 0.10 |
ENSDART00000131854
|
si:dkey-93m18.6
|
si:dkey-93m18.6 |
| chr15_-_43284021 | 0.10 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr8_-_30979494 | 0.10 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr7_+_21272833 | 0.09 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr20_+_6035427 | 0.09 |
ENSDART00000054086
|
tshr
|
thyroid stimulating hormone receptor |
| chr23_-_31969786 | 0.09 |
ENSDART00000134550
|
ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr24_-_40860603 | 0.09 |
ENSDART00000188032
|
CU633479.7
|
|
| chr17_+_51682429 | 0.09 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr11_+_17984354 | 0.09 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr6_-_40029423 | 0.09 |
ENSDART00000103230
|
pfkfb4b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4b |
| chr16_+_23303859 | 0.08 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr2_+_20406399 | 0.08 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr8_+_19504011 | 0.08 |
ENSDART00000158859
ENSDART00000159044 |
sec22bb
|
SEC22 homolog B, vesicle trafficking protein b |
| chr3_+_3681116 | 0.08 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr4_+_52212057 | 0.08 |
ENSDART00000165627
|
znf1034
|
zinc finger protein 1034 |
| chr23_+_4709607 | 0.08 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr17_-_10122204 | 0.08 |
ENSDART00000160751
|
BX088587.1
|
|
| chr25_+_18711804 | 0.08 |
ENSDART00000011149
|
fam185a
|
family with sequence similarity 185, member A |
| chr24_+_28953089 | 0.07 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr24_+_26328787 | 0.07 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr23_+_17509794 | 0.07 |
ENSDART00000148457
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr19_+_46158078 | 0.07 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr23_-_18567088 | 0.07 |
ENSDART00000192371
|
sephs2
|
selenophosphate synthetase 2 |
| chr24_+_26329018 | 0.07 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr16_+_13965923 | 0.07 |
ENSDART00000103857
|
zgc:162509
|
zgc:162509 |
| chr7_+_4474880 | 0.07 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr13_-_12602920 | 0.07 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr6_-_21582444 | 0.06 |
ENSDART00000151339
|
si:dkey-43k4.3
|
si:dkey-43k4.3 |
| chr17_+_22577472 | 0.06 |
ENSDART00000045099
|
yipf4
|
Yip1 domain family, member 4 |
| chr4_+_49266048 | 0.06 |
ENSDART00000127767
ENSDART00000186362 |
si:ch211-42i6.2
|
si:ch211-42i6.2 |
| chr13_+_33268657 | 0.05 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr17_-_25831569 | 0.05 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr8_-_12867128 | 0.05 |
ENSDART00000142201
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr21_-_13972745 | 0.05 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr22_-_28226948 | 0.05 |
ENSDART00000147686
|
si:dkey-222p3.1
|
si:dkey-222p3.1 |
| chr6_-_35052388 | 0.04 |
ENSDART00000181000
ENSDART00000170116 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr4_+_16885854 | 0.04 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr18_-_41161828 | 0.04 |
ENSDART00000114993
|
CABZ01005876.1
|
|
| chr23_+_4689626 | 0.04 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr13_-_9450210 | 0.04 |
ENSDART00000133768
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr11_-_40204621 | 0.04 |
ENSDART00000086287
|
znf362a
|
zinc finger protein 362a |
| chr8_-_53044300 | 0.03 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr17_+_45405821 | 0.03 |
ENSDART00000189482
ENSDART00000177422 |
tagapa
|
T cell activation RhoGTPase activating protein a |
| chr13_+_35528607 | 0.03 |
ENSDART00000075414
ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr8_+_43340995 | 0.03 |
ENSDART00000038566
|
rflna
|
refilin A |
| chr4_+_72723304 | 0.03 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr15_-_20939579 | 0.02 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr6_+_35052721 | 0.02 |
ENSDART00000191090
ENSDART00000082940 |
uhmk1
|
U2AF homology motif (UHM) kinase 1 |
| chr10_-_2524917 | 0.02 |
ENSDART00000188642
|
CU856539.1
|
|
| chr2_-_16217344 | 0.02 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr5_-_23843636 | 0.02 |
ENSDART00000193280
|
GBGT1 (1 of many)
|
si:ch211-135f11.5 |
| chr25_-_13490744 | 0.02 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr23_-_4409668 | 0.02 |
ENSDART00000081823
|
si:ch73-142c19.1
|
si:ch73-142c19.1 |
| chr10_-_11261386 | 0.02 |
ENSDART00000189946
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr11_+_13032483 | 0.01 |
ENSDART00000108968
|
ptgfr
|
prostaglandin F receptor (FP) |
| chr3_+_23029484 | 0.01 |
ENSDART00000187900
|
nags
|
N-acetylglutamate synthase |
| chr22_-_20812822 | 0.01 |
ENSDART00000193778
|
dot1l
|
DOT1-like histone H3K79 methyltransferase |
| chr6_+_17053650 | 0.01 |
ENSDART00000155296
|
pimr16
|
Pim proto-oncogene, serine/threonine kinase, related 16 |
| chr25_+_4855549 | 0.01 |
ENSDART00000163839
|
ap4e1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr1_+_51039558 | 0.01 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr7_+_27317174 | 0.01 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr25_+_22296666 | 0.01 |
ENSDART00000138887
ENSDART00000193410 |
cyp11a2
|
cytochrome P450, family 11, subfamily A, polypeptide 2 |
| chr13_+_646700 | 0.01 |
ENSDART00000006892
|
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr14_-_14607855 | 0.01 |
ENSDART00000162322
|
rab9b
|
RAB9B, member RAS oncogene family |
| chr14_-_451555 | 0.01 |
ENSDART00000190906
|
FAT4
|
FAT atypical cadherin 4 |
| chr16_+_26113457 | 0.00 |
ENSDART00000183537
|
BX571738.1
|
|
| chr14_-_413273 | 0.00 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr3_-_31619463 | 0.00 |
ENSDART00000124559
|
moto
|
minamoto |
| chr20_+_46202188 | 0.00 |
ENSDART00000100523
|
taar13c
|
trace amine associated receptor 13c |
| chr5_+_33498253 | 0.00 |
ENSDART00000140993
|
ms4a17c.2
|
membrane-spanning 4-domains, subfamily A, member 17c.2 |
| chr19_-_14970813 | 0.00 |
ENSDART00000184312
|
CR394532.2
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx1a+lhx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.2 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 0.3 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.3 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) |
| 0.0 | 0.1 | GO:0071498 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.5 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0016108 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.0 | 0.3 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.3 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.2 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.3 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.3 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.2 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.8 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |