Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
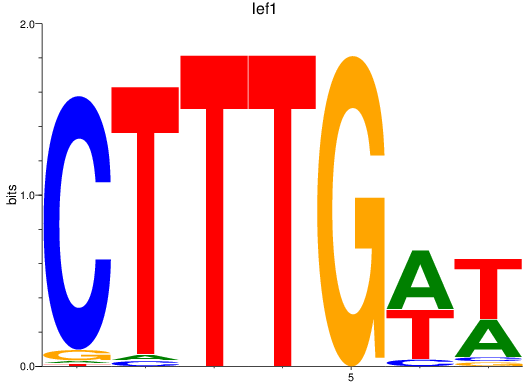
Results for lef1
Z-value: 2.12
Transcription factors associated with lef1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lef1
|
ENSDARG00000031894 | lymphoid enhancer-binding factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lef1 | dr11_v1_chr1_+_49814942_49814942 | -0.49 | 3.4e-02 | Click! |
Activity profile of lef1 motif
Sorted Z-values of lef1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_45705525 | 8.54 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr15_-_3736149 | 6.41 |
ENSDART00000182986
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr16_-_45910050 | 6.23 |
ENSDART00000133213
|
afp4
|
antifreeze protein type IV |
| chr12_+_6465557 | 6.23 |
ENSDART00000066477
ENSDART00000122271 |
dkk1b
|
dickkopf WNT signaling pathway inhibitor 1b |
| chr20_-_29498178 | 5.93 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr6_-_45905746 | 5.21 |
ENSDART00000025428
|
epha2a
|
eph receptor A2 a |
| chr19_-_17208728 | 4.73 |
ENSDART00000151228
|
stmn1a
|
stathmin 1a |
| chr18_-_23875370 | 4.62 |
ENSDART00000130163
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr6_-_43092175 | 4.50 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr17_+_15535501 | 4.20 |
ENSDART00000002932
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr9_+_15893093 | 4.08 |
ENSDART00000099483
ENSDART00000134657 |
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr23_-_3758637 | 4.06 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr3_+_41922114 | 3.96 |
ENSDART00000138280
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr21_-_37194365 | 3.86 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr24_-_12770357 | 3.85 |
ENSDART00000060826
|
ipo4
|
importin 4 |
| chr18_-_17513426 | 3.84 |
ENSDART00000146725
|
b3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr7_-_17337233 | 3.83 |
ENSDART00000050236
ENSDART00000102141 |
nitr8
|
novel immune-type receptor 8 |
| chr6_-_32093830 | 3.83 |
ENSDART00000017695
|
foxd3
|
forkhead box D3 |
| chr14_+_30543008 | 3.81 |
ENSDART00000145336
|
her5
|
hairy-related 5 |
| chr20_-_48485354 | 3.69 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr6_-_33913184 | 3.60 |
ENSDART00000146373
|
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr3_-_55650771 | 3.59 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr21_-_37194669 | 3.53 |
ENSDART00000192748
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr24_+_21514283 | 3.51 |
ENSDART00000007066
|
cdk8
|
cyclin-dependent kinase 8 |
| chr7_-_24112484 | 3.33 |
ENSDART00000111923
|
ajuba
|
ajuba LIM protein |
| chr14_+_6963312 | 3.29 |
ENSDART00000150050
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr17_+_38255105 | 3.15 |
ENSDART00000005296
|
nkx2.9
|
NK2 transcription factor related, locus 9 (Drosophila) |
| chr20_-_29499363 | 3.14 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr2_-_42960353 | 3.13 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr8_-_51753604 | 3.11 |
ENSDART00000007090
|
tbx16
|
T-box 16 |
| chr16_-_6198543 | 3.00 |
ENSDART00000167505
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr15_+_26933196 | 2.99 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr21_-_37027252 | 2.97 |
ENSDART00000076483
|
zgc:77151
|
zgc:77151 |
| chr19_+_19777437 | 2.96 |
ENSDART00000170662
|
hoxa3a
|
homeobox A3a |
| chr19_+_40026959 | 2.93 |
ENSDART00000123647
|
sfpq
|
splicing factor proline/glutamine-rich |
| chr3_-_26978793 | 2.88 |
ENSDART00000155396
|
nubp1
|
nucleotide binding protein 1 (MinD homolog, E. coli) |
| chr16_-_45917322 | 2.87 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr8_-_22964486 | 2.86 |
ENSDART00000112381
|
emilin3a
|
elastin microfibril interfacer 3a |
| chr1_+_8694196 | 2.86 |
ENSDART00000025604
|
zgc:77849
|
zgc:77849 |
| chr5_+_67662430 | 2.85 |
ENSDART00000137700
ENSDART00000142586 |
si:dkey-70b23.2
|
si:dkey-70b23.2 |
| chr9_-_33877476 | 2.81 |
ENSDART00000150035
ENSDART00000088441 ENSDART00000183210 |
si:ch73-147f11.1
|
si:ch73-147f11.1 |
| chr3_-_55650417 | 2.81 |
ENSDART00000171441
|
axin2
|
axin 2 (conductin, axil) |
| chr21_-_23305561 | 2.80 |
ENSDART00000181815
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr14_-_25930182 | 2.74 |
ENSDART00000018651
ENSDART00000147991 |
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr9_+_25568839 | 2.72 |
ENSDART00000177342
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr1_+_10110203 | 2.64 |
ENSDART00000080576
ENSDART00000181437 |
lratb.1
|
lecithin retinol acyltransferase b, tandem duplicate 1 |
| chr5_+_42280372 | 2.64 |
ENSDART00000142855
|
tbx6l
|
T-box 6, like |
| chr5_+_10014604 | 2.63 |
ENSDART00000092333
|
slc2a11b
|
solute carrier family 2 (facilitated glucose transporter), member 11b |
| chr1_-_36772147 | 2.62 |
ENSDART00000053369
|
prmt9
|
protein arginine methyltransferase 9 |
| chr15_-_3736773 | 2.62 |
ENSDART00000090624
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr22_+_13917311 | 2.61 |
ENSDART00000022654
|
sh3bp4a
|
SH3-domain binding protein 4a |
| chr5_+_49744713 | 2.61 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr10_+_32104305 | 2.60 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr5_+_27442247 | 2.60 |
ENSDART00000184129
|
loxl2b
|
lysyl oxidase-like 2b |
| chr25_+_34576067 | 2.59 |
ENSDART00000157519
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr14_-_33981544 | 2.59 |
ENSDART00000167774
|
foxa
|
forkhead box A sequence |
| chr2_+_11760548 | 2.57 |
ENSDART00000057268
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr11_-_23219367 | 2.56 |
ENSDART00000003646
|
optc
|
opticin |
| chr8_+_19668654 | 2.55 |
ENSDART00000091436
|
foxe3
|
forkhead box E3 |
| chr11_-_17755444 | 2.55 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr21_-_28920245 | 2.54 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr21_-_20328375 | 2.53 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr3_+_23721808 | 2.49 |
ENSDART00000012470
|
hoxb4a
|
homeobox B4a |
| chr19_+_43523690 | 2.49 |
ENSDART00000113031
|
wasf2
|
WAS protein family, member 2 |
| chr17_+_35243753 | 2.48 |
ENSDART00000016702
|
iah1
|
isoamyl acetate hydrolyzing esterase 1 (putative) |
| chr11_-_30138299 | 2.48 |
ENSDART00000172106
|
scml2
|
Scm polycomb group protein like 2 |
| chr2_-_49031303 | 2.47 |
ENSDART00000143471
|
cdc34b
|
cell division cycle 34 homolog (S. cerevisiae) b |
| chr21_-_20321660 | 2.46 |
ENSDART00000188497
|
zgc:86764
|
zgc:86764 |
| chr10_-_29892486 | 2.44 |
ENSDART00000099983
|
bsx
|
brain-specific homeobox |
| chr5_+_60590796 | 2.44 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr2_+_37838259 | 2.40 |
ENSDART00000136796
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr5_-_24517768 | 2.39 |
ENSDART00000003957
|
tia1l
|
cytotoxic granule-associated RNA binding protein 1, like |
| chr13_+_5978809 | 2.39 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr24_+_4978055 | 2.37 |
ENSDART00000045813
|
zic4
|
zic family member 4 |
| chr14_+_7932973 | 2.34 |
ENSDART00000109941
|
cxxc5b
|
CXXC finger protein 5b |
| chr5_-_24231139 | 2.34 |
ENSDART00000143492
|
senp3a
|
SUMO1/sentrin/SMT3 specific peptidase 3a |
| chr14_+_7699443 | 2.32 |
ENSDART00000123139
|
brd8
|
bromodomain containing 8 |
| chr8_-_16697912 | 2.29 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr18_+_24922125 | 2.29 |
ENSDART00000180385
|
rgma
|
repulsive guidance molecule family member a |
| chr22_-_10541372 | 2.29 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr7_-_54677143 | 2.28 |
ENSDART00000163748
|
ccnd1
|
cyclin D1 |
| chr14_+_6962271 | 2.27 |
ENSDART00000148447
ENSDART00000149114 ENSDART00000149492 ENSDART00000148394 |
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr18_-_23875219 | 2.26 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr23_+_31245395 | 2.25 |
ENSDART00000053588
|
irak1bp1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr7_-_17028015 | 2.25 |
ENSDART00000022441
|
dbx1a
|
developing brain homeobox 1a |
| chr19_-_47570672 | 2.24 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr24_+_4977862 | 2.24 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr4_+_12031958 | 2.23 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr8_-_1264893 | 2.23 |
ENSDART00000190371
|
cdc14b
|
cell division cycle 14B |
| chr22_-_16154771 | 2.19 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr5_-_66028371 | 2.19 |
ENSDART00000183012
|
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr2_+_30423428 | 2.19 |
ENSDART00000139871
|
socs6b
|
suppressor of cytokine signaling 6b |
| chr22_-_18546241 | 2.19 |
ENSDART00000105404
ENSDART00000105405 |
cirbpb
|
cold inducible RNA binding protein b |
| chr13_-_35808904 | 2.17 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr23_-_22072716 | 2.16 |
ENSDART00000054362
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr8_-_41228530 | 2.15 |
ENSDART00000165949
ENSDART00000173055 |
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr20_+_4221978 | 2.14 |
ENSDART00000171898
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr2_-_30668580 | 2.12 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr9_+_6802641 | 2.11 |
ENSDART00000187278
|
FO203514.1
|
|
| chr11_+_6116503 | 2.10 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr4_-_27117112 | 2.09 |
ENSDART00000034534
|
zbed4
|
zinc finger, BED-type containing 4 |
| chr11_+_30296332 | 2.09 |
ENSDART00000192843
|
ugt1b7
|
UDP glucuronosyltransferase 1 family, polypeptide B7 |
| chr9_+_34148714 | 2.09 |
ENSDART00000078051
|
gpr161
|
G protein-coupled receptor 161 |
| chr9_-_27410597 | 2.09 |
ENSDART00000135652
ENSDART00000042297 |
kdelc1
|
KDEL (Lys-Asp-Glu-Leu) containing 1 |
| chr4_-_11580948 | 2.08 |
ENSDART00000049066
|
net1
|
neuroepithelial cell transforming 1 |
| chr24_+_14451404 | 2.07 |
ENSDART00000123674
ENSDART00000066720 |
prdm14
|
PR domain containing 14 |
| chr6_-_10728057 | 2.06 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr16_-_16237844 | 2.04 |
ENSDART00000168747
ENSDART00000111912 |
rbm12b
|
RNA binding motif protein 12B |
| chr12_+_27231212 | 2.04 |
ENSDART00000133023
ENSDART00000123739 |
tmem106a
|
transmembrane protein 106A |
| chr11_+_31609481 | 2.04 |
ENSDART00000124830
ENSDART00000162768 |
zgc:162816
|
zgc:162816 |
| chr4_-_2945306 | 2.02 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr10_-_17466990 | 2.02 |
ENSDART00000147794
|
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr3_-_25377163 | 2.01 |
ENSDART00000055490
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr3_+_26245731 | 2.00 |
ENSDART00000103734
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr22_+_5120033 | 1.99 |
ENSDART00000169200
|
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr6_+_296130 | 1.98 |
ENSDART00000073985
|
rbfox2
|
RNA binding fox-1 homolog 2 |
| chr23_-_10175898 | 1.97 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr6_-_55297274 | 1.97 |
ENSDART00000184283
|
ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr25_+_18953575 | 1.95 |
ENSDART00000155610
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr18_-_19819812 | 1.93 |
ENSDART00000060344
|
aagab
|
alpha and gamma adaptin binding protein |
| chr2_+_29976419 | 1.93 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr1_-_9130136 | 1.93 |
ENSDART00000113013
|
palb2
|
partner and localizer of BRCA2 |
| chr22_-_26005894 | 1.92 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr25_+_18964782 | 1.91 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr22_-_10541712 | 1.90 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr22_+_16497670 | 1.90 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr18_-_17087138 | 1.89 |
ENSDART00000135597
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr6_+_43426599 | 1.89 |
ENSDART00000056457
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr21_-_18275226 | 1.89 |
ENSDART00000126672
ENSDART00000135239 |
brd3a
|
bromodomain containing 3a |
| chr17_-_45383699 | 1.89 |
ENSDART00000141182
|
tmem206
|
transmembrane protein 206 |
| chr25_+_3294150 | 1.88 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr25_+_31323978 | 1.88 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr6_+_6780873 | 1.88 |
ENSDART00000011865
|
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr8_+_17869225 | 1.88 |
ENSDART00000080079
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr17_-_42213285 | 1.87 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr22_+_18349794 | 1.87 |
ENSDART00000186580
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr5_-_65021156 | 1.87 |
ENSDART00000166183
|
anxa1c
|
annexin A1c |
| chr11_+_33818179 | 1.87 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr8_-_50287949 | 1.87 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr22_+_11153590 | 1.85 |
ENSDART00000188207
|
bcor
|
BCL6 corepressor |
| chr3_+_15809098 | 1.85 |
ENSDART00000183023
|
phospho1
|
phosphatase, orphan 1 |
| chr2_+_38271392 | 1.83 |
ENSDART00000042100
|
homeza
|
homeobox and leucine zipper encoding a |
| chr6_-_8377055 | 1.83 |
ENSDART00000131513
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr19_-_30420602 | 1.83 |
ENSDART00000144121
|
ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr1_+_36772348 | 1.81 |
ENSDART00000109314
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr20_-_3319642 | 1.81 |
ENSDART00000186743
ENSDART00000123096 |
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr19_+_7424347 | 1.81 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr9_-_45601103 | 1.81 |
ENSDART00000180465
|
agr1
|
anterior gradient 1 |
| chr2_-_28102264 | 1.81 |
ENSDART00000013638
|
cdh6
|
cadherin 6 |
| chr5_-_65000312 | 1.79 |
ENSDART00000192893
|
ANXA1 (1 of many)
|
zgc:110283 |
| chr12_+_33919502 | 1.79 |
ENSDART00000085888
|
trim8b
|
tripartite motif containing 8b |
| chr20_-_43723860 | 1.78 |
ENSDART00000122051
|
mixl1
|
Mix paired-like homeobox |
| chr19_-_19720744 | 1.78 |
ENSDART00000170636
|
evx1
|
even-skipped homeobox 1 |
| chr17_-_2595736 | 1.78 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr20_-_22484621 | 1.78 |
ENSDART00000063601
|
gsx2
|
GS homeobox 2 |
| chr9_-_7683799 | 1.78 |
ENSDART00000102713
|
si:ch73-199e17.1
|
si:ch73-199e17.1 |
| chr19_-_14191592 | 1.77 |
ENSDART00000164594
|
tbxta
|
T-box transcription factor Ta |
| chr10_+_17714866 | 1.77 |
ENSDART00000039969
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr23_+_36144487 | 1.77 |
ENSDART00000082473
|
hoxc3a
|
homeobox C3a |
| chr19_-_19721556 | 1.77 |
ENSDART00000165196
|
evx1
|
even-skipped homeobox 1 |
| chr6_+_15762647 | 1.77 |
ENSDART00000127133
ENSDART00000128939 |
iqca1
|
IQ motif containing with AAA domain 1 |
| chr23_+_36122058 | 1.76 |
ENSDART00000184448
|
hoxc3a
|
homeobox C3a |
| chr9_-_34507680 | 1.76 |
ENSDART00000113656
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr14_+_25505468 | 1.76 |
ENSDART00000079016
|
thoc3
|
THO complex 3 |
| chr6_+_54711306 | 1.75 |
ENSDART00000074605
|
pkp1b
|
plakophilin 1b |
| chr13_+_10023256 | 1.74 |
ENSDART00000110035
|
srbd1
|
S1 RNA binding domain 1 |
| chr5_+_25762271 | 1.74 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr4_+_12612145 | 1.74 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr13_-_45022301 | 1.72 |
ENSDART00000183589
ENSDART00000125633 ENSDART00000074787 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr7_+_38811800 | 1.72 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr7_-_51648749 | 1.70 |
ENSDART00000109055
|
cited1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr16_-_29194517 | 1.70 |
ENSDART00000046114
ENSDART00000148899 |
mef2d
|
myocyte enhancer factor 2d |
| chr3_+_34988670 | 1.69 |
ENSDART00000011319
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr5_-_41531629 | 1.69 |
ENSDART00000051082
|
akr1a1a
|
aldo-keto reductase family 1, member A1a (aldehyde reductase) |
| chr11_-_18705303 | 1.68 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr14_-_39031108 | 1.68 |
ENSDART00000026194
|
glra4a
|
glycine receptor, alpha 4a |
| chr7_+_24006875 | 1.68 |
ENSDART00000033755
|
homezb
|
homeobox and leucine zipper encoding b |
| chr7_-_26270014 | 1.68 |
ENSDART00000079347
|
serpine1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr8_+_26034623 | 1.68 |
ENSDART00000004521
ENSDART00000142555 |
arih2
|
ariadne homolog 2 (Drosophila) |
| chr14_+_32022272 | 1.68 |
ENSDART00000105760
|
zic6
|
zic family member 6 |
| chr4_-_21466480 | 1.68 |
ENSDART00000139962
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr6_+_43400059 | 1.67 |
ENSDART00000143374
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr25_+_17849223 | 1.66 |
ENSDART00000144484
|
PYURF
|
zgc:162634 |
| chr8_+_20488322 | 1.66 |
ENSDART00000036630
|
zgc:101100
|
zgc:101100 |
| chr14_+_22498757 | 1.66 |
ENSDART00000021657
|
smyd5
|
SMYD family member 5 |
| chr24_+_35911300 | 1.65 |
ENSDART00000129679
|
abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr9_-_12401871 | 1.65 |
ENSDART00000191901
|
nup35
|
nucleoporin 35 |
| chr7_+_20260172 | 1.65 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr2_+_30481125 | 1.64 |
ENSDART00000125933
|
fam173b
|
family with sequence similarity 173, member B |
| chr5_-_22082918 | 1.64 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr3_-_52661242 | 1.63 |
ENSDART00000138018
|
zgc:113210
|
zgc:113210 |
| chr2_+_26179096 | 1.62 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr7_+_20019125 | 1.62 |
ENSDART00000186391
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr12_+_27231607 | 1.62 |
ENSDART00000066270
|
tmem106a
|
transmembrane protein 106A |
| chr11_-_14813029 | 1.62 |
ENSDART00000004920
ENSDART00000122645 |
sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr9_-_7684002 | 1.61 |
ENSDART00000016360
|
si:ch73-199e17.1
|
si:ch73-199e17.1 |
| chr1_+_38153944 | 1.61 |
ENSDART00000135666
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr10_+_9550419 | 1.61 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr13_+_29771463 | 1.61 |
ENSDART00000134424
ENSDART00000138332 ENSDART00000134330 ENSDART00000160944 ENSDART00000076992 ENSDART00000160921 |
pax2a
|
paired box 2a |
| chr13_-_35908275 | 1.61 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
Network of associatons between targets according to the STRING database.
First level regulatory network of lef1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 2.5 | 7.4 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 1.2 | 11.2 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 1.2 | 8.5 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 1.2 | 4.9 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 1.1 | 3.3 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 1.0 | 11.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 1.0 | 3.0 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 1.0 | 3.9 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.9 | 2.8 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.9 | 3.6 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.9 | 3.5 | GO:0016109 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.9 | 3.5 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.9 | 2.6 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) |
| 0.8 | 3.9 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.8 | 3.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.7 | 6.0 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.7 | 2.0 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.7 | 2.0 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.7 | 4.0 | GO:0055016 | hypochord development(GO:0055016) |
| 0.7 | 4.6 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.7 | 2.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.6 | 2.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.6 | 3.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.6 | 0.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.6 | 3.5 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.6 | 2.3 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.6 | 2.3 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.6 | 4.5 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.5 | 1.6 | GO:1903644 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.5 | 2.1 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.5 | 1.5 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.5 | 2.9 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.5 | 2.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.4 | 14.0 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.4 | 5.2 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.4 | 1.3 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.4 | 3.0 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.4 | 4.2 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.4 | 1.7 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.4 | 4.2 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.4 | 1.2 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.4 | 2.0 | GO:0036088 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.4 | 1.9 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.4 | 1.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.4 | 5.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.4 | 1.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.4 | 1.9 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.4 | 1.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.4 | 3.3 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.4 | 1.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.4 | 5.0 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.3 | 2.4 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.3 | 0.3 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.3 | 1.3 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.3 | 3.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.3 | 2.3 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.3 | 1.3 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.3 | 1.3 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.3 | 0.9 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.3 | 1.2 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.3 | 3.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.3 | 2.4 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.3 | 2.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.3 | 2.7 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.3 | 3.9 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.3 | 0.9 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.3 | 2.6 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.3 | 2.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.3 | 4.7 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.3 | 1.7 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 | 3.8 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 11.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 | 1.1 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.3 | 3.6 | GO:1901797 | negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.3 | 0.8 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 2.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.2 | 1.7 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 0.7 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.2 | 0.7 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.2 | 2.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 | 0.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 1.2 | GO:0072104 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.2 | 3.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 2.5 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 0.7 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.2 | 3.8 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.2 | 1.1 | GO:0060584 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 | 1.5 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.2 | 2.8 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.2 | 1.5 | GO:0090169 | regulation of spindle assembly(GO:0090169) regulation of mitotic spindle assembly(GO:1901673) |
| 0.2 | 0.6 | GO:1903060 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.2 | 1.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 0.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 2.6 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.2 | 1.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 2.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 2.5 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.2 | 0.6 | GO:0030237 | female sex determination(GO:0030237) |
| 0.2 | 0.6 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.2 | 0.6 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.2 | 1.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 1.6 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 0.5 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.2 | 0.9 | GO:0022029 | telencephalon cell migration(GO:0022029) |
| 0.2 | 0.7 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.2 | 0.7 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.2 | 0.9 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 | 2.6 | GO:2000826 | regulation of heart morphogenesis(GO:2000826) |
| 0.2 | 1.0 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.2 | 0.5 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 2.9 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.2 | 1.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 | 1.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.2 | 1.7 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.2 | 0.6 | GO:0035909 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.2 | 0.5 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.2 | 1.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 0.6 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 1.1 | GO:0019857 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.2 | 2.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.7 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.2 | 0.8 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 1.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 1.0 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.7 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 2.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 2.3 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 3.9 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.1 | 1.8 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 3.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 1.9 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 1.0 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 2.0 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.0 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 2.9 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.6 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 2.8 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.5 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.6 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.5 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.1 | 0.5 | GO:0003190 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.1 | 1.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.5 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 0.6 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.1 | 1.9 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 1.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.7 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 5.3 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 0.3 | GO:0061181 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) regulation of chondrocyte development(GO:0061181) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.1 | 2.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.2 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.7 | GO:0014846 | vascular endothelial growth factor production(GO:0010573) regulation of vascular endothelial growth factor production(GO:0010574) positive regulation of vascular endothelial growth factor production(GO:0010575) esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.3 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.1 | 1.9 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 1.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.2 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.1 | 1.1 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.2 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.1 | 0.5 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 1.7 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.1 | 2.9 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 1.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.1 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.1 | 0.3 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.7 | GO:0072422 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.1 | 0.3 | GO:0042256 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.9 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 1.3 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.1 | 5.0 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.1 | 0.3 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.1 | 2.2 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 1.5 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.1 | 0.2 | GO:0003097 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) |
| 0.1 | 0.5 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 1.4 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.1 | 0.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.7 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 0.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 1.7 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 1.5 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 2.5 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 1.4 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.3 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.1 | 0.4 | GO:0045822 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.1 | 1.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 0.6 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 3.0 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 2.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.2 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.1 | 0.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.2 | GO:0010893 | positive regulation of steroid biosynthetic process(GO:0010893) |
| 0.1 | 0.8 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.1 | 2.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 3.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.2 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 1.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 2.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 0.3 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 1.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 1.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.7 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.8 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 2.5 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.1 | 1.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.6 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.1 | 2.4 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 0.8 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.1 | 0.6 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 2.1 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 0.3 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.1 | 1.5 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 5.1 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 0.8 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 0.3 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.6 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.9 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 1.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.4 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 6.2 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.1 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.4 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.1 | 1.2 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 1.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.7 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.1 | 0.7 | GO:2000027 | regulation of organ morphogenesis(GO:2000027) |
| 0.1 | 1.8 | GO:0061053 | somite development(GO:0061053) |
| 0.1 | 0.5 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.7 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 2.6 | GO:0001764 | neuron migration(GO:0001764) |
| 0.1 | 1.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 2.6 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.1 | 1.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.5 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 1.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.9 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.7 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.5 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.0 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.8 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 1.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.3 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.3 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 1.1 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.9 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 1.4 | GO:0039021 | pronephric glomerulus development(GO:0039021) |
| 0.0 | 0.3 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 1.8 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 0.5 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 1.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 1.1 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.7 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.5 | GO:0034394 | protein localization to cell surface(GO:0034394) energy homeostasis(GO:0097009) |
| 0.0 | 1.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.3 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.0 | 0.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 1.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling(GO:0098917) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.0 | 0.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 9.7 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 7.0 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 3.1 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 2.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 8.4 | GO:0045892 | negative regulation of transcription, DNA-templated(GO:0045892) |
| 0.0 | 0.5 | GO:0070816 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.9 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.3 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.7 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 2.5 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 4.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.5 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.9 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.4 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 1.3 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 2.8 | GO:0090504 | epiboly(GO:0090504) |
| 0.0 | 1.2 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 1.8 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.0 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.4 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 2.3 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 1.0 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 0.4 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 0.9 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.2 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.2 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.1 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.4 | GO:0021551 | central nervous system morphogenesis(GO:0021551) protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.3 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.0 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.4 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.5 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 1.7 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.2 | GO:0045762 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.2 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 1.1 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 4.1 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.5 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 1.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.5 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.5 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.7 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.5 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.5 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:0002685 | regulation of leukocyte migration(GO:0002685) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.6 | 1.8 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.5 | 1.6 | GO:0072380 | TRC complex(GO:0072380) |
| 0.5 | 3.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.4 | 10.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.4 | 1.2 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.4 | 2.9 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 1.3 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.3 | 0.9 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.3 | 4.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.3 | 2.9 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 9.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 3.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 0.9 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 0.6 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.2 | 3.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 4.0 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.2 | 1.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 3.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 1.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 2.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 0.6 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 1.7 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 2.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 0.7 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 1.2 | GO:0031464 | inclusion body(GO:0016234) Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 2.4 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 0.7 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 1.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 2.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 0.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 3.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 0.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 3.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 1.4 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.9 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.5 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 1.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 2.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.0 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.5 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 0.8 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 1.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 2.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.1 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 1.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 2.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.7 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 2.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 1.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 2.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 2.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 4.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 7.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 1.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 4.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 4.3 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 3.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 4.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 1.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 3.6 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.5 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.1 | 1.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.5 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 1.0 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 3.1 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 11.4 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 2.3 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.1 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 3.3 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.6 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 112.1 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 8.2 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 1.3 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 9.1 | GO:0005829 | cytosol(GO:0005829) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 11.3 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.2 | 6.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 1.2 | 8.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 1.2 | 9.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 1.0 | 4.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.9 | 3.5 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.7 | 9.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.7 | 2.0 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.6 | 1.8 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.6 | 5.2 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.6 | 2.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.6 | 2.3 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.6 | 7.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 1.5 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.5 | 4.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.5 | 2.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.4 | 4.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.4 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 2.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.4 | 1.6 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.4 | 2.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.4 | 1.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.4 | 1.8 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.3 | 5.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 4.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.3 | 5.9 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.3 | 1.7 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.3 | 3.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 2.6 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.3 | 0.9 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.3 | 1.9 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.3 | 1.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 1.1 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 4.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.3 | 2.0 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.3 | 1.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.3 | 2.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 3.8 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.3 | 2.7 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.3 | 4.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 0.7 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.2 | 0.7 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.2 | 0.9 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 1.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 3.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 1.1 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.2 | 4.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.2 | 5.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 0.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 1.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 1.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 1.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 2.4 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.2 | 1.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 0.8 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 1.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 1.7 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 1.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 1.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 0.5 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.2 | 0.7 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.2 | 0.7 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.2 | 1.2 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.2 | 2.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 1.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 1.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.2 | 2.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 5.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 2.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 0.7 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 2.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.4 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.1 | 3.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.5 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 1.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.5 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 4.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 2.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 3.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.5 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.1 | 0.6 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 0.3 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 1.4 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 0.4 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.7 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.3 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 2.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 7.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.6 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 0.8 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 4.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 1.5 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 14.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 3.2 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 1.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 4.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 2.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 4.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 3.5 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 2.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.3 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.7 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.2 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 2.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.8 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 1.1 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 4.0 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 2.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 1.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 1.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 4.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 4.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 7.8 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 4.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.7 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 1.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 68.2 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 5.6 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 2.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.2 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 1.1 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.7 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 15.2 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 2.6 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.3 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 2.0 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.0 | 15.4 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.1 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.5 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 1.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.0 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.7 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 4.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.0 | GO:0003676 | nucleic acid binding(GO:0003676) |
| 0.0 | 0.2 | GO:0015926 | glucosidase activity(GO:0015926) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.3 | 4.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 3.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 3.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 13.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 15.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 2.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 2.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 2.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 2.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.7 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 1.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 1.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 2.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 2.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 2.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 0.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 2.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.4 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 2.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.3 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.8 | 7.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.6 | 12.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 2.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 4.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 2.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 6.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 5.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 1.6 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 10.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 5.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 2.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 4.8 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 0.6 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 2.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 3.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 5.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 2.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.8 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.7 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 1.6 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.2 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 2.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |