Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
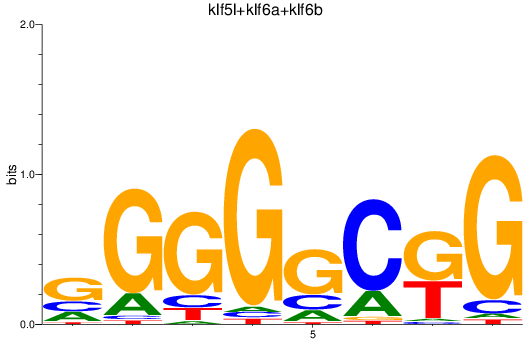
Results for klf5l+klf6a+klf6b
Z-value: 0.46
Transcription factors associated with klf5l+klf6a+klf6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf5l
|
ENSDARG00000018757 | Kruppel-like factor 5 like |
|
klf6a
|
ENSDARG00000029072 | Kruppel-like factor 6a |
|
klf6b
|
ENSDARG00000038561 | Kruppel-like factor 6b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf6b | dr11_v1_chr2_+_48074243_48074243 | 0.72 | 4.9e-04 | Click! |
| klf6a | dr11_v1_chr24_-_4148914_4148915 | -0.45 | 5.6e-02 | Click! |
| klf5l | dr11_v1_chr21_-_38153824_38153824 | 0.10 | 7.0e-01 | Click! |
Activity profile of klf5l+klf6a+klf6b motif
Sorted Z-values of klf5l+klf6a+klf6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_44026369 | 1.72 |
ENSDART00000185456
|
crybb1
|
crystallin, beta B1 |
| chr8_+_43056153 | 1.64 |
ENSDART00000186377
|
prnpa
|
prion protein a |
| chr20_+_9355912 | 1.29 |
ENSDART00000142337
|
si:dkey-174m14.3
|
si:dkey-174m14.3 |
| chr3_+_32526263 | 1.27 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr6_-_13891727 | 1.13 |
ENSDART00000065356
|
desmb
|
desmin b |
| chr6_-_60104628 | 1.06 |
ENSDART00000057463
ENSDART00000169188 |
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr24_+_36452927 | 0.70 |
ENSDART00000168181
|
zgc:114120
|
zgc:114120 |
| chr18_+_660578 | 0.69 |
ENSDART00000161203
|
CELF6
|
si:dkey-205h23.2 |
| chr3_+_1223824 | 0.66 |
ENSDART00000065922
|
wbp2nl
|
WBP2 N-terminal like |
| chr15_-_25613114 | 0.65 |
ENSDART00000182431
ENSDART00000187405 |
taok1a
|
TAO kinase 1a |
| chr20_-_7176809 | 0.65 |
ENSDART00000012247
ENSDART00000125019 |
dhcr24
|
24-dehydrocholesterol reductase |
| chr21_+_45733871 | 0.63 |
ENSDART00000187285
ENSDART00000193018 |
zgc:77058
|
zgc:77058 |
| chr20_+_4392687 | 0.63 |
ENSDART00000187271
|
im:7142702
|
im:7142702 |
| chr2_+_59015878 | 0.59 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr2_-_32501501 | 0.57 |
ENSDART00000181309
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr2_+_24867534 | 0.54 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr22_+_12366516 | 0.51 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr25_+_2229301 | 0.50 |
ENSDART00000180198
|
pparab
|
peroxisome proliferator-activated receptor alpha b |
| chr3_-_62380146 | 0.49 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr21_+_43253538 | 0.49 |
ENSDART00000179940
ENSDART00000164806 ENSDART00000147026 |
shroom1
|
shroom family member 1 |
| chr24_-_28381404 | 0.47 |
ENSDART00000148406
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr21_-_24865454 | 0.44 |
ENSDART00000142907
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr15_-_47956388 | 0.43 |
ENSDART00000116506
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr2_+_24868010 | 0.40 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr8_-_4100365 | 0.39 |
ENSDART00000142846
|
cux2b
|
cut-like homeobox 2b |
| chr15_+_20543770 | 0.37 |
ENSDART00000092357
|
sgsm2
|
small G protein signaling modulator 2 |
| chr1_-_10647484 | 0.37 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr22_+_1006573 | 0.34 |
ENSDART00000123458
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr4_+_3438510 | 0.34 |
ENSDART00000155320
|
atxn7l1
|
ataxin 7-like 1 |
| chr20_-_1268863 | 0.33 |
ENSDART00000109321
ENSDART00000027119 |
lats1
|
large tumor suppressor kinase 1 |
| chr21_-_37733571 | 0.33 |
ENSDART00000176214
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr13_+_23093743 | 0.30 |
ENSDART00000148034
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr12_-_47648538 | 0.29 |
ENSDART00000108477
|
fh
|
fumarate hydratase |
| chr21_-_24865217 | 0.29 |
ENSDART00000101136
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr2_+_33335911 | 0.28 |
ENSDART00000126135
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr4_+_57093908 | 0.28 |
ENSDART00000170198
|
si:ch211-238e22.5
|
si:ch211-238e22.5 |
| chr15_-_45538773 | 0.27 |
ENSDART00000113494
|
MB21D2
|
Mab-21 domain containing 2 |
| chr10_-_38456382 | 0.25 |
ENSDART00000182129
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr10_+_21650828 | 0.25 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr9_+_44722205 | 0.25 |
ENSDART00000086176
ENSDART00000145271 ENSDART00000132696 |
nckap1
|
NCK-associated protein 1 |
| chr21_+_6613772 | 0.25 |
ENSDART00000159645
|
col5a1
|
procollagen, type V, alpha 1 |
| chr10_+_41549819 | 0.24 |
ENSDART00000114210
|
CABZ01049847.1
|
|
| chr9_+_17787864 | 0.24 |
ENSDART00000013111
|
dgkh
|
diacylglycerol kinase, eta |
| chr13_-_50200042 | 0.23 |
ENSDART00000074230
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr16_-_563235 | 0.23 |
ENSDART00000016303
|
irx2a
|
iroquois homeobox 2a |
| chr25_-_6629281 | 0.22 |
ENSDART00000112782
|
cspg4
|
chondroitin sulfate proteoglycan 4 |
| chr17_+_37253706 | 0.22 |
ENSDART00000076004
|
tmem62
|
transmembrane protein 62 |
| chr1_-_10647307 | 0.21 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr13_-_7767548 | 0.20 |
ENSDART00000158720
ENSDART00000121952 |
h2afy2
|
H2A histone family, member Y2 |
| chr8_+_36621598 | 0.20 |
ENSDART00000180389
|
magixb
|
MAGI family member, X-linked b |
| chr23_+_42338325 | 0.19 |
ENSDART00000169660
|
cyp2aa7
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 |
| chr13_+_33688474 | 0.18 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr22_+_396840 | 0.18 |
ENSDART00000163198
|
capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr9_+_13999620 | 0.17 |
ENSDART00000143229
|
cd28l
|
cd28-like molecule |
| chr19_-_20106486 | 0.17 |
ENSDART00000043924
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr20_-_49889111 | 0.16 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr21_-_5056812 | 0.16 |
ENSDART00000139713
ENSDART00000140859 |
zgc:77838
|
zgc:77838 |
| chr21_-_37733287 | 0.16 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr4_+_53119675 | 0.16 |
ENSDART00000167470
|
si:dkey-8o9.2
|
si:dkey-8o9.2 |
| chr17_+_6793001 | 0.15 |
ENSDART00000030773
|
foxo3a
|
forkhead box O3A |
| chr8_+_8298439 | 0.14 |
ENSDART00000170566
|
srpk3
|
SRSF protein kinase 3 |
| chr9_+_38427572 | 0.14 |
ENSDART00000108860
|
CYP27A1 (1 of many)
|
zgc:136333 |
| chr22_+_344763 | 0.13 |
ENSDART00000181934
|
CU914780.1
|
|
| chr9_-_8454060 | 0.10 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr22_+_35089031 | 0.10 |
ENSDART00000076040
|
srfa
|
serum response factor a |
| chr22_-_26558166 | 0.10 |
ENSDART00000111125
|
glis2a
|
GLIS family zinc finger 2a |
| chr23_-_524780 | 0.09 |
ENSDART00000055139
|
col9a3
|
collagen, type IX, alpha 3 |
| chr6_+_12503849 | 0.07 |
ENSDART00000149529
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr3_-_30909487 | 0.05 |
ENSDART00000025046
|
ppp1caa
|
protein phosphatase 1, catalytic subunit, alpha isozyme a |
| chr15_-_38202630 | 0.04 |
ENSDART00000183772
|
rhoga
|
ras homolog family member Ga |
| chr7_-_23768234 | 0.04 |
ENSDART00000173981
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr5_-_37252111 | 0.03 |
ENSDART00000185110
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr6_-_10788065 | 0.02 |
ENSDART00000190968
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr25_+_27493444 | 0.01 |
ENSDART00000112299
|
gpr37a
|
G protein-coupled receptor 37a |
| chr22_+_30496360 | 0.01 |
ENSDART00000189145
|
BX649490.4
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of klf5l+klf6a+klf6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.2 | 0.5 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.2 | 1.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.7 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 1.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.3 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.0 | 0.3 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.7 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.2 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.3 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.3 | GO:0003139 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
| 0.0 | 0.6 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.2 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 1.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.0 | 0.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 1.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 1.6 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |