Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
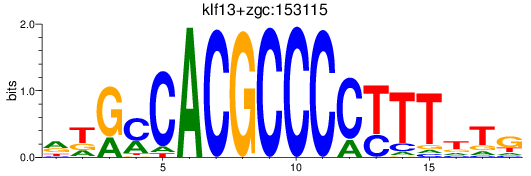
Results for klf13+zgc:153115
Z-value: 0.54
Transcription factors associated with klf13+zgc:153115
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf13
|
ENSDARG00000061368 | Kruppel-like factor 13 |
|
zgc
|
ENSDARG00000069342 | 153115 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf13 | dr11_v1_chr25_-_34512102_34512102 | 0.17 | 5.0e-01 | Click! |
| zgc:153115 | dr11_v1_chr2_-_7246848_7246848 | 0.08 | 7.5e-01 | Click! |
Activity profile of klf13+zgc:153115 motif
Sorted Z-values of klf13+zgc:153115 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_69482891 | 1.63 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr12_+_49125510 | 1.61 |
ENSDART00000185804
|
FO704607.1
|
|
| chr23_-_43393083 | 0.98 |
ENSDART00000112758
|
zgc:174862
|
zgc:174862 |
| chr23_+_28693278 | 0.96 |
ENSDART00000078148
|
smc1a
|
structural maintenance of chromosomes 1A |
| chr11_-_4235811 | 0.85 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr11_+_1796426 | 0.78 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr16_+_10329701 | 0.68 |
ENSDART00000172845
|
mdc1
|
mediator of DNA damage checkpoint 1 |
| chr1_-_12109216 | 0.64 |
ENSDART00000079930
|
mttp
|
microsomal triglyceride transfer protein |
| chr5_+_11812089 | 0.64 |
ENSDART00000111359
|
fbxo21
|
F-box protein 21 |
| chr5_+_68807170 | 0.64 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr12_+_30046320 | 0.63 |
ENSDART00000179904
ENSDART00000153394 |
ablim1b
|
actin binding LIM protein 1b |
| chr22_-_6562618 | 0.59 |
ENSDART00000106100
|
zgc:171490
|
zgc:171490 |
| chr1_-_55196103 | 0.58 |
ENSDART00000140153
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr1_+_9004719 | 0.57 |
ENSDART00000006211
ENSDART00000137211 |
prkcba
|
protein kinase C, beta a |
| chr7_-_5396154 | 0.53 |
ENSDART00000172980
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr10_+_1638876 | 0.53 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr10_+_4875262 | 0.52 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr18_+_33725576 | 0.52 |
ENSDART00000146816
|
si:dkey-145c18.5
|
si:dkey-145c18.5 |
| chr6_+_54576520 | 0.50 |
ENSDART00000093199
ENSDART00000127519 ENSDART00000157142 |
tead3b
|
TEA domain family member 3 b |
| chr22_-_6801876 | 0.50 |
ENSDART00000135726
|
si:ch1073-188e1.1
|
si:ch1073-188e1.1 |
| chr4_-_22519516 | 0.48 |
ENSDART00000130409
ENSDART00000186258 ENSDART00000002851 ENSDART00000123801 |
kdm7aa
|
lysine (K)-specific demethylase 7Aa |
| chr17_-_4318393 | 0.47 |
ENSDART00000167995
ENSDART00000153824 |
napba
|
N-ethylmaleimide-sensitive factor attachment protein, beta a |
| chr1_-_52201266 | 0.46 |
ENSDART00000143805
ENSDART00000023757 |
rab3da
|
RAB3D, member RAS oncogene family, a |
| chr8_-_53490376 | 0.45 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr23_-_45398622 | 0.43 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr16_-_1757521 | 0.41 |
ENSDART00000124660
|
ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr12_-_13556536 | 0.40 |
ENSDART00000166396
|
CR354395.1
|
|
| chr15_+_45544589 | 0.40 |
ENSDART00000055978
|
LO018197.1
|
|
| chr16_+_43077909 | 0.40 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
| chr7_+_72460911 | 0.39 |
ENSDART00000160682
ENSDART00000168532 |
HECTD4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr6_-_54111928 | 0.39 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr17_+_389218 | 0.39 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr19_+_46113828 | 0.39 |
ENSDART00000159331
ENSDART00000161826 |
rbm24a
|
RNA binding motif protein 24a |
| chr10_+_6383270 | 0.39 |
ENSDART00000170548
|
zgc:114200
|
zgc:114200 |
| chr3_-_40054615 | 0.38 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr22_-_6777976 | 0.37 |
ENSDART00000187946
|
CT583625.5
|
|
| chr8_+_10862353 | 0.37 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr24_-_39633224 | 0.36 |
ENSDART00000105646
|
fam234a
|
family with sequence similarity 234, member A |
| chr17_-_34963575 | 0.36 |
ENSDART00000145664
|
kidins220a
|
kinase D-interacting substrate 220a |
| chr5_-_30924347 | 0.35 |
ENSDART00000111749
ENSDART00000086564 ENSDART00000153909 |
spns2
|
spinster homolog 2 (Drosophila) |
| chr25_-_20268027 | 0.34 |
ENSDART00000138763
|
dnajb9a
|
DnaJ (Hsp40) homolog, subfamily B, member 9a |
| chr8_+_29963024 | 0.34 |
ENSDART00000149372
|
ptch1
|
patched 1 |
| chr5_+_67812062 | 0.34 |
ENSDART00000158611
|
zgc:175280
|
zgc:175280 |
| chr21_+_6197223 | 0.33 |
ENSDART00000147716
|
si:dkey-93m18.3
|
si:dkey-93m18.3 |
| chr6_+_16406723 | 0.33 |
ENSDART00000040035
|
ccdc80l1
|
coiled-coil domain containing 80 like 1 |
| chr9_-_37749973 | 0.32 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr10_-_1625080 | 0.32 |
ENSDART00000137285
|
nup155
|
nucleoporin 155 |
| chr6_+_3282809 | 0.31 |
ENSDART00000187444
ENSDART00000187407 ENSDART00000191883 |
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr22_-_16275236 | 0.30 |
ENSDART00000149051
|
cdc14ab
|
cell division cycle 14Ab |
| chr3_+_19299309 | 0.30 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr19_-_205104 | 0.30 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr3_+_31925067 | 0.29 |
ENSDART00000127330
ENSDART00000055279 |
snrnp70
|
small nuclear ribonucleoprotein 70 (U1) |
| chr21_+_6114709 | 0.29 |
ENSDART00000065858
|
fpgs
|
folylpolyglutamate synthase |
| chr13_-_46421682 | 0.29 |
ENSDART00000149602
|
fgfr2
|
fibroblast growth factor receptor 2 |
| chr3_+_59117136 | 0.29 |
ENSDART00000182745
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr22_-_6968756 | 0.29 |
ENSDART00000136636
|
fgfr1bl
|
fibroblast growth factor receptor 1b, like |
| chr2_+_55365727 | 0.28 |
ENSDART00000162943
|
FP245456.1
|
|
| chr4_+_76492691 | 0.28 |
ENSDART00000186337
|
CU104731.1
|
|
| chr19_+_9305964 | 0.27 |
ENSDART00000136241
|
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr3_-_47876427 | 0.27 |
ENSDART00000180844
ENSDART00000124480 |
adgrl1a
|
adhesion G protein-coupled receptor L1a |
| chr21_+_13182149 | 0.27 |
ENSDART00000140267
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr7_-_73846995 | 0.27 |
ENSDART00000188079
|
FP236812.4
|
|
| chr19_-_2421793 | 0.27 |
ENSDART00000180238
|
TMEM196 (1 of many)
|
transmembrane protein 196 |
| chr21_+_6114305 | 0.27 |
ENSDART00000141607
|
fpgs
|
folylpolyglutamate synthase |
| chr12_+_2676878 | 0.26 |
ENSDART00000185909
|
antxr1c
|
anthrax toxin receptor 1c |
| chr1_-_34335752 | 0.26 |
ENSDART00000140157
|
si:dkey-24h22.5
|
si:dkey-24h22.5 |
| chr10_+_41668483 | 0.26 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr17_+_8323348 | 0.26 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr14_+_36869273 | 0.25 |
ENSDART00000111674
|
F2RL3
|
si:ch211-132p1.2 |
| chr24_+_17005647 | 0.25 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr10_+_38643304 | 0.25 |
ENSDART00000067447
|
mmp30
|
matrix metallopeptidase 30 |
| chr10_+_22034477 | 0.24 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr10_-_4980150 | 0.24 |
ENSDART00000093228
|
mat2al
|
methionine adenosyltransferase II, alpha-like |
| chr1_-_34450784 | 0.24 |
ENSDART00000140515
|
lmo7b
|
LIM domain 7b |
| chr16_-_13613475 | 0.24 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr8_-_13046089 | 0.23 |
ENSDART00000137784
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr8_-_52143109 | 0.23 |
ENSDART00000142969
|
tcf7l1b
|
transcription factor 7 like 1b |
| chr20_-_36617313 | 0.23 |
ENSDART00000172395
ENSDART00000152856 |
enah
|
enabled homolog (Drosophila) |
| chr6_-_18976168 | 0.22 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr8_+_47219107 | 0.22 |
ENSDART00000146018
ENSDART00000075068 |
mthfr
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr7_+_1473929 | 0.22 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr9_-_27649406 | 0.22 |
ENSDART00000181270
ENSDART00000187112 |
stxbp5l
|
syntaxin binding protein 5-like |
| chr2_-_48153945 | 0.22 |
ENSDART00000146553
|
pfkpb
|
phosphofructokinase, platelet b |
| chr10_+_3299829 | 0.22 |
ENSDART00000183684
|
zgc:56235
|
zgc:56235 |
| chr8_+_29962635 | 0.22 |
ENSDART00000007640
|
ptch1
|
patched 1 |
| chr17_+_2162916 | 0.22 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr11_-_25733910 | 0.21 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr18_+_6558338 | 0.21 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr18_-_6943577 | 0.21 |
ENSDART00000132399
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr10_-_41980797 | 0.20 |
ENSDART00000076575
|
rhof
|
ras homolog family member F |
| chr22_+_2495929 | 0.20 |
ENSDART00000178351
|
CU856622.1
|
|
| chr23_-_43393589 | 0.19 |
ENSDART00000183592
|
zgc:174862
|
zgc:174862 |
| chr18_+_6558146 | 0.19 |
ENSDART00000169401
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr1_+_604127 | 0.19 |
ENSDART00000133165
|
jam2a
|
junctional adhesion molecule 2a |
| chr10_+_1668106 | 0.19 |
ENSDART00000142278
|
sgsm1b
|
small G protein signaling modulator 1b |
| chr6_+_7414215 | 0.19 |
ENSDART00000049339
|
sox21a
|
SRY (sex determining region Y)-box 21a |
| chr1_+_7189856 | 0.19 |
ENSDART00000092114
|
si:ch73-383l1.1
|
si:ch73-383l1.1 |
| chr3_+_634682 | 0.19 |
ENSDART00000164305
|
dicp1.1
|
diverse immunoglobulin domain-containing protein 1.1 |
| chr25_-_4235037 | 0.19 |
ENSDART00000093003
|
syt7a
|
synaptotagmin VIIa |
| chr17_+_51764310 | 0.19 |
ENSDART00000157171
|
si:ch211-168d23.3
|
si:ch211-168d23.3 |
| chr24_-_12745222 | 0.19 |
ENSDART00000151836
|
si:ch211-196f5.9
|
si:ch211-196f5.9 |
| chr8_-_21268303 | 0.18 |
ENSDART00000067211
|
gpr37l1b
|
G protein-coupled receptor 37 like 1b |
| chr19_+_22727940 | 0.18 |
ENSDART00000052509
|
trhrb
|
thyrotropin-releasing hormone receptor b |
| chr18_+_507835 | 0.17 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr8_+_104114 | 0.17 |
ENSDART00000172101
|
sncaip
|
synuclein, alpha interacting protein |
| chr17_-_37795865 | 0.17 |
ENSDART00000025853
|
zfp36l1a
|
zinc finger protein 36, C3H type-like 1a |
| chr13_-_42673978 | 0.17 |
ENSDART00000133848
ENSDART00000099738 ENSDART00000099729 ENSDART00000169083 |
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr15_-_46828958 | 0.17 |
ENSDART00000045110
|
chrd
|
chordin |
| chr23_-_35483163 | 0.17 |
ENSDART00000138660
ENSDART00000113643 ENSDART00000189269 |
fbxo25
|
F-box protein 25 |
| chr7_+_20260172 | 0.16 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr7_+_58730201 | 0.16 |
ENSDART00000073640
|
plag1
|
pleiomorphic adenoma gene 1 |
| chr10_-_10864331 | 0.16 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr8_+_52637507 | 0.16 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr9_+_4378153 | 0.15 |
ENSDART00000191264
ENSDART00000182384 |
kalrna
|
kalirin RhoGEF kinase a |
| chr7_-_6592142 | 0.15 |
ENSDART00000160137
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr9_-_19489264 | 0.15 |
ENSDART00000122894
|
wdr4
|
WD repeat domain 4 |
| chr15_-_5563551 | 0.15 |
ENSDART00000099520
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr11_+_11152214 | 0.15 |
ENSDART00000148030
|
ly75
|
lymphocyte antigen 75 |
| chr21_-_41070182 | 0.14 |
ENSDART00000026064
|
larsb
|
leucyl-tRNA synthetase b |
| chr21_-_11657043 | 0.14 |
ENSDART00000141297
|
cast
|
calpastatin |
| chr21_+_28535203 | 0.14 |
ENSDART00000184950
|
CR749775.2
|
|
| chr5_+_61657702 | 0.13 |
ENSDART00000134387
ENSDART00000171248 |
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr1_+_45839927 | 0.13 |
ENSDART00000148086
ENSDART00000180413 ENSDART00000048191 ENSDART00000179047 |
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr5_+_43458304 | 0.13 |
ENSDART00000051114
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr3_+_5575313 | 0.12 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr19_-_15397946 | 0.12 |
ENSDART00000143480
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr3_-_27915270 | 0.12 |
ENSDART00000115370
|
mettl22
|
methyltransferase like 22 |
| chr24_-_33780387 | 0.12 |
ENSDART00000079210
|
cdk5
|
cyclin-dependent kinase 5 |
| chr21_-_3201027 | 0.12 |
ENSDART00000161256
|
zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr21_+_43854594 | 0.12 |
ENSDART00000142718
|
csf1rb
|
colony stimulating factor 1 receptor, b |
| chr17_-_45386823 | 0.12 |
ENSDART00000156002
|
tmem206
|
transmembrane protein 206 |
| chr15_+_32821392 | 0.12 |
ENSDART00000158272
|
dclk1b
|
doublecortin-like kinase 1b |
| chr7_+_52887701 | 0.11 |
ENSDART00000109973
|
tp53bp1
|
tumor protein p53 binding protein, 1 |
| chr10_+_44940693 | 0.11 |
ENSDART00000157515
|
cnnm4a
|
cyclin and CBS domain divalent metal cation transport mediator 4a |
| chr23_+_44157682 | 0.11 |
ENSDART00000164474
ENSDART00000149928 |
si:ch73-106g13.1
|
si:ch73-106g13.1 |
| chr16_-_9425451 | 0.11 |
ENSDART00000149163
|
ccr8.1
|
chemokine (C-C motif) receptor 8.1 |
| chr20_-_25748407 | 0.11 |
ENSDART00000063152
|
chst14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 |
| chr20_+_23501535 | 0.11 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr17_-_45387134 | 0.11 |
ENSDART00000010975
|
tmem206
|
transmembrane protein 206 |
| chr10_-_5821584 | 0.10 |
ENSDART00000166388
|
il6st
|
interleukin 6 signal transducer |
| chr4_-_39448829 | 0.10 |
ENSDART00000168526
|
si:dkey-261o4.9
|
si:dkey-261o4.9 |
| chr1_+_56755536 | 0.10 |
ENSDART00000157727
|
CABZ01049361.1
|
|
| chr19_-_32944050 | 0.09 |
ENSDART00000137611
|
azin1b
|
antizyme inhibitor 1b |
| chr4_+_9400012 | 0.09 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr24_-_36238054 | 0.09 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr7_-_64152087 | 0.09 |
ENSDART00000165234
|
CR354398.1
|
|
| chr24_-_37688398 | 0.09 |
ENSDART00000141414
|
zgc:112185
|
zgc:112185 |
| chr14_-_24101897 | 0.09 |
ENSDART00000143695
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr7_+_52364748 | 0.09 |
ENSDART00000006795
|
gnrhr2
|
gonadotropin releasing hormone receptor 2 |
| chr18_+_27077853 | 0.09 |
ENSDART00000125326
ENSDART00000192660 ENSDART00000098334 |
ppp1r15b
|
protein phosphatase 1, regulatory subunit 15B |
| chr23_-_790620 | 0.09 |
ENSDART00000166818
|
CU927920.1
|
|
| chr6_-_43233700 | 0.09 |
ENSDART00000113706
|
trnt1
|
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr21_+_37436907 | 0.09 |
ENSDART00000182611
ENSDART00000076328 |
pgrmc1
|
progesterone receptor membrane component 1 |
| chr21_+_45386033 | 0.09 |
ENSDART00000151773
|
jade2
|
jade family PHD finger 2 |
| chr15_-_3078600 | 0.09 |
ENSDART00000172842
|
fndc3a
|
fibronectin type III domain containing 3A |
| chr7_+_69528850 | 0.09 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr16_-_26132122 | 0.08 |
ENSDART00000157787
|
lipeb
|
lipase, hormone-sensitive b |
| chr7_-_69647988 | 0.08 |
ENSDART00000169943
|
LO018231.1
|
|
| chr12_-_3053873 | 0.08 |
ENSDART00000023796
ENSDART00000137148 |
dcxr
|
dicarbonyl/L-xylulose reductase |
| chr13_+_13033424 | 0.08 |
ENSDART00000159441
|
letm1
|
leucine zipper-EF-hand containing transmembrane protein 1 |
| chr16_-_42523744 | 0.08 |
ENSDART00000017185
|
tbx20
|
T-box 20 |
| chr1_-_34450622 | 0.08 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr3_-_16039619 | 0.08 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr19_-_3878548 | 0.08 |
ENSDART00000168377
ENSDART00000172271 |
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr3_+_36284986 | 0.07 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr11_+_13071645 | 0.07 |
ENSDART00000162259
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr16_+_39271123 | 0.07 |
ENSDART00000043823
ENSDART00000141801 |
osbpl10b
|
oxysterol binding protein-like 10b |
| chr23_+_10396842 | 0.07 |
ENSDART00000167593
ENSDART00000125103 |
eif4ba
|
eukaryotic translation initiation factor 4Ba |
| chr10_-_44017642 | 0.07 |
ENSDART00000135240
ENSDART00000014669 |
acads
|
acyl-CoA dehydrogenase short chain |
| chr6_-_10034145 | 0.07 |
ENSDART00000185999
|
nudt15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr4_+_9679840 | 0.07 |
ENSDART00000190889
|
BX901962.12
|
|
| chr18_-_14677936 | 0.07 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr13_-_27916439 | 0.07 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr17_-_51260476 | 0.06 |
ENSDART00000084348
|
trappc12
|
trafficking protein particle complex 12 |
| chr19_+_30495109 | 0.06 |
ENSDART00000192229
|
BX510301.1
|
|
| chr5_+_13549603 | 0.06 |
ENSDART00000171270
|
ckap2l
|
cytoskeleton associated protein 2-like |
| chr4_-_5455506 | 0.06 |
ENSDART00000156593
ENSDART00000154676 |
si:dkey-14d8.22
|
si:dkey-14d8.22 |
| chr7_-_3470451 | 0.06 |
ENSDART00000173387
|
si:ch211-285c6.6
|
si:ch211-285c6.6 |
| chr5_-_72136548 | 0.06 |
ENSDART00000007827
|
spra
|
sepiapterin reductase a |
| chr12_-_3053699 | 0.06 |
ENSDART00000139721
|
dcxr
|
dicarbonyl/L-xylulose reductase |
| chr25_+_34581494 | 0.06 |
ENSDART00000167690
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr1_-_493218 | 0.06 |
ENSDART00000031635
|
ercc5
|
excision repair cross-complementation group 5 |
| chr18_+_507618 | 0.06 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr15_+_28989573 | 0.05 |
ENSDART00000076648
|
clip3
|
CAP-GLY domain containing linker protein 3 |
| chr23_-_41821825 | 0.05 |
ENSDART00000184725
|
GMEB2
|
si:ch73-302a13.2 |
| chr10_+_10728870 | 0.05 |
ENSDART00000109282
|
swi5
|
SWI5 homologous recombination repair protein |
| chr15_-_1001177 | 0.05 |
ENSDART00000160730
|
zgc:162936
|
zgc:162936 |
| chr9_+_907459 | 0.05 |
ENSDART00000034850
ENSDART00000144114 |
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr20_+_46311707 | 0.04 |
ENSDART00000184743
|
flvcr2b
|
feline leukemia virus subgroup C cellular receptor family, member 2b |
| chr3_+_5297493 | 0.04 |
ENSDART00000138596
|
si:ch211-150d5.3
|
si:ch211-150d5.3 |
| chr21_-_13230925 | 0.04 |
ENSDART00000023834
|
setb
|
SET nuclear proto-oncogene b |
| chr22_-_38224315 | 0.04 |
ENSDART00000165430
ENSDART00000140968 |
hps3
|
Hermansky-Pudlak syndrome 3 |
| chr6_-_43233373 | 0.04 |
ENSDART00000188945
|
trnt1
|
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr25_+_3507368 | 0.04 |
ENSDART00000157777
|
zgc:153293
|
zgc:153293 |
| chr6_-_17849786 | 0.04 |
ENSDART00000172709
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr22_+_2217939 | 0.04 |
ENSDART00000145394
ENSDART00000146694 |
znf1172
|
zinc finger protein 1172 |
| chr8_-_4100365 | 0.03 |
ENSDART00000142846
|
cux2b
|
cut-like homeobox 2b |
| chr3_-_33934788 | 0.03 |
ENSDART00000151411
|
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr20_+_572037 | 0.03 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr14_+_97017 | 0.02 |
ENSDART00000159300
ENSDART00000169523 |
mcm7
|
minichromosome maintenance complex component 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of klf13+zgc:153115
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.2 | 0.5 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.6 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.4 | GO:2001014 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.3 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.6 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.6 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 0.3 | GO:0097638 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.4 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.3 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.2 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.3 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 1.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.2 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.2 | GO:0039689 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.1 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.1 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.4 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.6 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0061620 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0060043 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.1 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.5 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.2 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 0.1 | GO:0097037 | heme export(GO:0097037) |
| 0.0 | 0.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.1 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.5 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 1.0 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.6 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 0.6 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.4 | GO:1990825 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.5 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.4 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.4 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 0.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.3 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.8 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |