Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
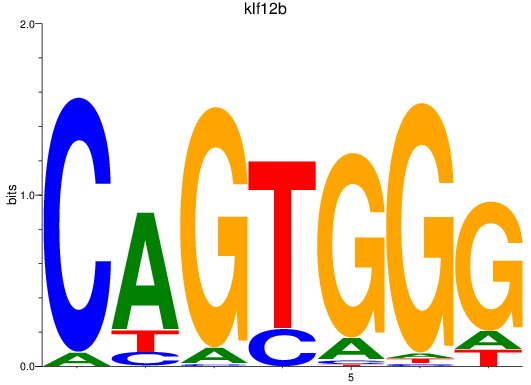
Results for klf12b
Z-value: 1.20
Transcription factors associated with klf12b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf12b
|
ENSDARG00000032197 | Kruppel-like factor 12b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf12b | dr11_v1_chr9_+_30720048_30720048 | 0.42 | 7.7e-02 | Click! |
Activity profile of klf12b motif
Sorted Z-values of klf12b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25187210 | 5.49 |
ENSDART00000101147
ENSDART00000167528 |
si:dkey-183i3.5
|
si:dkey-183i3.5 |
| chr6_+_13742899 | 4.51 |
ENSDART00000104722
|
cdk5r2a
|
cyclin-dependent kinase 5, regulatory subunit 2a (p39) |
| chr19_-_26863626 | 3.73 |
ENSDART00000145568
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr15_-_37846047 | 3.56 |
ENSDART00000184837
|
hsc70
|
heat shock cognate 70 |
| chr5_-_25174420 | 2.41 |
ENSDART00000141554
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr22_-_282498 | 2.36 |
ENSDART00000182766
|
CABZ01079178.1
|
|
| chr5_-_67471375 | 2.30 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr13_-_11536951 | 2.15 |
ENSDART00000018155
|
adss
|
adenylosuccinate synthase |
| chr10_-_19497914 | 2.15 |
ENSDART00000132084
|
si:ch211-127i16.2
|
si:ch211-127i16.2 |
| chr11_+_36989696 | 2.13 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr4_-_1324141 | 2.09 |
ENSDART00000180720
|
ptn
|
pleiotrophin |
| chr13_-_31166544 | 2.07 |
ENSDART00000146250
ENSDART00000132129 ENSDART00000139591 |
mapk8a
|
mitogen-activated protein kinase 8a |
| chr8_-_37263524 | 2.07 |
ENSDART00000061327
|
rh50
|
Rh50-like protein |
| chr5_-_22501663 | 2.00 |
ENSDART00000133174
|
si:dkey-27p18.5
|
si:dkey-27p18.5 |
| chr18_-_46241775 | 1.99 |
ENSDART00000145999
ENSDART00000134244 ENSDART00000185021 ENSDART00000181855 |
prx
|
periaxin |
| chr9_-_35155089 | 1.94 |
ENSDART00000077901
|
appb
|
amyloid beta (A4) precursor protein b |
| chr22_-_263117 | 1.90 |
ENSDART00000158134
|
zgc:66156
|
zgc:66156 |
| chr2_+_28672152 | 1.89 |
ENSDART00000157410
ENSDART00000169614 |
nadsyn1
|
NAD synthetase 1 |
| chr22_+_7439186 | 1.82 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr2_+_54696042 | 1.82 |
ENSDART00000074270
|
ankrd12
|
ankyrin repeat domain 12 |
| chr2_+_24203229 | 1.81 |
ENSDART00000138088
|
map4l
|
microtubule associated protein 4 like |
| chr17_+_18117358 | 1.75 |
ENSDART00000144894
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr10_+_5135842 | 1.74 |
ENSDART00000132627
ENSDART00000162434 |
zgc:113274
|
zgc:113274 |
| chr11_-_37509001 | 1.72 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr4_-_68563862 | 1.68 |
ENSDART00000182970
|
BX548011.2
|
|
| chr1_-_44434707 | 1.66 |
ENSDART00000110148
|
cryba1l2
|
crystallin, beta A1, like 2 |
| chr8_-_14050758 | 1.66 |
ENSDART00000133922
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr11_+_41135055 | 1.66 |
ENSDART00000173252
|
camta1
|
calmodulin binding transcription activator 1 |
| chr19_+_164013 | 1.64 |
ENSDART00000167379
|
carmil1
|
capping protein regulator and myosin 1 linker 1 |
| chr1_+_14283692 | 1.61 |
ENSDART00000017679
|
ppp2r2ca
|
protein phosphatase 2, regulatory subunit B, gamma a |
| chr20_+_5564042 | 1.56 |
ENSDART00000090934
ENSDART00000127050 |
nrxn3b
|
neurexin 3b |
| chr11_+_23957440 | 1.54 |
ENSDART00000190721
|
cntn2
|
contactin 2 |
| chr23_-_39636195 | 1.54 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr21_-_28235361 | 1.51 |
ENSDART00000164458
|
nrxn2a
|
neurexin 2a |
| chr20_+_6142433 | 1.50 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr25_+_12640211 | 1.50 |
ENSDART00000165108
|
jph3
|
junctophilin 3 |
| chr17_-_15546862 | 1.49 |
ENSDART00000091021
|
col10a1a
|
collagen, type X, alpha 1a |
| chr8_-_25247284 | 1.47 |
ENSDART00000132697
|
gnat2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr4_+_6643421 | 1.45 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr5_-_68927728 | 1.44 |
ENSDART00000132838
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr25_-_4146947 | 1.43 |
ENSDART00000129268
|
fads2
|
fatty acid desaturase 2 |
| chr7_-_24520866 | 1.43 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr22_-_294700 | 1.43 |
ENSDART00000189179
|
CABZ01079179.1
|
|
| chr14_-_8890437 | 1.42 |
ENSDART00000167242
|
ATG2A
|
si:ch73-45o6.2 |
| chr2_+_47582681 | 1.42 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr1_-_51465730 | 1.41 |
ENSDART00000074284
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr8_-_53108207 | 1.41 |
ENSDART00000111023
|
b3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
| chr7_+_21841037 | 1.40 |
ENSDART00000077503
|
tm4sf5
|
transmembrane 4 L six family member 5 |
| chr10_+_22381802 | 1.37 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr15_+_37197494 | 1.37 |
ENSDART00000166203
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr10_-_20637098 | 1.37 |
ENSDART00000080391
|
sprn2
|
shadow of prion protein 2 |
| chr10_-_22828302 | 1.36 |
ENSDART00000079459
ENSDART00000100468 |
per1a
|
period circadian clock 1a |
| chr23_+_16633951 | 1.35 |
ENSDART00000109537
ENSDART00000193323 |
snphb
|
syntaphilin b |
| chr19_+_37458610 | 1.33 |
ENSDART00000103151
|
dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr23_-_24483311 | 1.33 |
ENSDART00000185793
ENSDART00000109248 |
spen
|
spen family transcriptional repressor |
| chr6_-_39006449 | 1.31 |
ENSDART00000150885
|
vdrb
|
vitamin D receptor b |
| chr21_+_6290566 | 1.31 |
ENSDART00000161647
|
fnbp1b
|
formin binding protein 1b |
| chr23_+_10352921 | 1.31 |
ENSDART00000081193
|
KRT18 (1 of many)
|
si:ch211-133j6.3 |
| chr22_-_600016 | 1.30 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr16_-_12173554 | 1.29 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr15_-_12319065 | 1.28 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr24_+_40529346 | 1.25 |
ENSDART00000168548
|
CU929259.1
|
|
| chr10_-_14929392 | 1.25 |
ENSDART00000137430
|
smad2
|
SMAD family member 2 |
| chr23_+_44883805 | 1.24 |
ENSDART00000182805
|
si:ch73-361h17.1
|
si:ch73-361h17.1 |
| chr13_+_25853757 | 1.23 |
ENSDART00000132521
|
bcl11aa
|
B cell CLL/lymphoma 11Aa |
| chr5_-_23317477 | 1.22 |
ENSDART00000090171
|
nlgn3b
|
neuroligin 3b |
| chr5_-_13076779 | 1.22 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr20_+_19119800 | 1.22 |
ENSDART00000151881
|
rp1l1b
|
retinitis pigmentosa 1-like 1b |
| chr1_-_22512063 | 1.20 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr24_+_29381946 | 1.20 |
ENSDART00000189551
|
ntng1a
|
netrin g1a |
| chr2_+_22694382 | 1.19 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr21_-_43949208 | 1.18 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr19_-_44089509 | 1.18 |
ENSDART00000189136
|
rad21b
|
RAD21 cohesin complex component b |
| chr21_+_6291027 | 1.17 |
ENSDART00000180467
ENSDART00000184952 ENSDART00000184006 |
fnbp1b
|
formin binding protein 1b |
| chr4_+_23223881 | 1.16 |
ENSDART00000133056
ENSDART00000089126 |
trhde.1
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 1 |
| chr22_-_22130623 | 1.14 |
ENSDART00000113168
|
CU855878.1
|
|
| chr5_-_38155005 | 1.14 |
ENSDART00000097770
|
gucy2d
|
guanylate cyclase 2D, retinal |
| chr4_-_77557279 | 1.13 |
ENSDART00000180113
|
AL935186.10
|
|
| chr16_-_27749172 | 1.13 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr21_+_10739846 | 1.12 |
ENSDART00000084011
|
cplx4a
|
complexin 4a |
| chr14_+_28281744 | 1.12 |
ENSDART00000173292
|
mid2
|
midline 2 |
| chr9_+_49659114 | 1.11 |
ENSDART00000189643
|
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr7_+_36539124 | 1.10 |
ENSDART00000173653
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr7_+_24573721 | 1.10 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr12_+_24344963 | 1.09 |
ENSDART00000191648
ENSDART00000183180 ENSDART00000088178 ENSDART00000189696 |
nrxn1a
|
neurexin 1a |
| chr18_+_18104235 | 1.08 |
ENSDART00000145342
|
cbln1
|
cerebellin 1 precursor |
| chr6_-_30210378 | 1.07 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr4_-_23643272 | 1.07 |
ENSDART00000112301
ENSDART00000133184 |
trhde.2
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 2 |
| chr22_+_7486867 | 1.07 |
ENSDART00000034586
|
CELA1 (1 of many)
|
zgc:112302 |
| chr8_+_28900689 | 1.06 |
ENSDART00000141634
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr6_+_40591149 | 1.05 |
ENSDART00000189060
ENSDART00000188298 |
frs3
|
fibroblast growth factor receptor substrate 3 |
| chr24_+_29382109 | 1.05 |
ENSDART00000184620
ENSDART00000188414 ENSDART00000186132 ENSDART00000191489 |
ntng1a
|
netrin g1a |
| chr11_+_23933016 | 1.04 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr10_+_20180163 | 1.04 |
ENSDART00000080016
|
ppp3ccb
|
protein phosphatase 3, catalytic subunit, gamma isozyme, b |
| chr18_-_37007294 | 1.03 |
ENSDART00000088309
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr10_+_29698467 | 1.03 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr15_+_24212847 | 1.03 |
ENSDART00000155502
|
sez6b
|
seizure related 6 homolog b |
| chr13_+_28702104 | 1.02 |
ENSDART00000135481
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr18_-_15771551 | 1.02 |
ENSDART00000130931
ENSDART00000154079 |
si:ch211-219a15.3
|
si:ch211-219a15.3 |
| chr6_-_39158953 | 1.01 |
ENSDART00000050682
|
stat2
|
signal transducer and activator of transcription 2 |
| chr14_-_2209742 | 1.01 |
ENSDART00000054889
|
pcdh2ab5
|
protocadherin 2 alpha b 5 |
| chr24_+_39137001 | 1.01 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr6_+_24817852 | 1.01 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr14_-_2213660 | 1.00 |
ENSDART00000162537
|
pcdh2ab3
|
protocadherin 2 alpha b 3 |
| chr1_-_12160981 | 1.00 |
ENSDART00000162146
|
b4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr6_-_7842078 | 0.99 |
ENSDART00000065507
|
plppr2b
|
phospholipid phosphatase related 2b |
| chr5_+_20148671 | 0.99 |
ENSDART00000143205
|
svopa
|
SV2 related protein a |
| chr19_-_13733870 | 0.98 |
ENSDART00000177773
|
epb41a
|
erythrocyte membrane protein band 4.1a |
| chr4_+_20263097 | 0.98 |
ENSDART00000138820
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr25_+_19955598 | 0.98 |
ENSDART00000091547
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr11_-_27874116 | 0.98 |
ENSDART00000180579
|
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr17_-_51829310 | 0.98 |
ENSDART00000154544
|
numb
|
numb homolog (Drosophila) |
| chr20_-_31781941 | 0.97 |
ENSDART00000139417
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr18_-_37007061 | 0.96 |
ENSDART00000136432
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr23_-_8373676 | 0.96 |
ENSDART00000105135
ENSDART00000158531 |
oprl1
|
opiate receptor-like 1 |
| chr4_+_19535946 | 0.96 |
ENSDART00000192342
ENSDART00000183740 ENSDART00000180812 ENSDART00000180017 |
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr9_+_54290896 | 0.95 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr16_-_44512882 | 0.95 |
ENSDART00000191241
|
CR925804.2
|
|
| chr16_-_40727455 | 0.95 |
ENSDART00000162331
|
si:dkey-22o22.2
|
si:dkey-22o22.2 |
| chr17_+_43623598 | 0.94 |
ENSDART00000154138
|
znf365
|
zinc finger protein 365 |
| chr2_+_9822319 | 0.94 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr3_-_40955780 | 0.94 |
ENSDART00000130130
|
cyp3c3
|
cytochrome P450, family 3, subfamily c, polypeptide 3 |
| chr21_-_12272543 | 0.94 |
ENSDART00000081510
ENSDART00000151297 |
celf4
|
CUGBP, Elav-like family member 4 |
| chr15_-_9593532 | 0.93 |
ENSDART00000169912
|
gab2
|
GRB2-associated binding protein 2 |
| chr5_-_21970881 | 0.93 |
ENSDART00000182907
|
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr23_+_19590598 | 0.93 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr16_+_6944311 | 0.93 |
ENSDART00000144763
|
eaf1
|
ELL associated factor 1 |
| chr22_-_10482253 | 0.93 |
ENSDART00000143164
|
aspn
|
asporin (LRR class 1) |
| chr20_+_1412193 | 0.93 |
ENSDART00000064419
|
leg1.1
|
liver-enriched gene 1, tandem duplicate 1 |
| chr14_-_14607855 | 0.92 |
ENSDART00000162322
|
rab9b
|
RAB9B, member RAS oncogene family |
| chr9_-_32604414 | 0.92 |
ENSDART00000088876
ENSDART00000166502 |
satb2
|
SATB homeobox 2 |
| chr23_+_16638639 | 0.92 |
ENSDART00000143545
|
snphb
|
syntaphilin b |
| chr21_+_21195487 | 0.91 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr24_-_11325849 | 0.90 |
ENSDART00000182485
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr15_-_26686908 | 0.90 |
ENSDART00000185254
|
pitpnm3
|
PITPNM family member 3 |
| chr18_-_12612699 | 0.90 |
ENSDART00000090335
|
hipk2
|
homeodomain interacting protein kinase 2 |
| chr13_-_5937034 | 0.90 |
ENSDART00000045292
|
spred2b
|
sprouty-related, EVH1 domain containing 2b |
| chr17_-_36896560 | 0.89 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr7_-_57933736 | 0.89 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr20_-_1378514 | 0.88 |
ENSDART00000181830
|
scara5
|
scavenger receptor class A, member 5 (putative) |
| chr25_-_19090479 | 0.88 |
ENSDART00000027465
ENSDART00000177670 |
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr12_+_16233077 | 0.87 |
ENSDART00000152409
|
mpp3b
|
membrane protein, palmitoylated 3b (MAGUK p55 subfamily member 3) |
| chr18_-_16179129 | 0.87 |
ENSDART00000125353
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr3_+_23247325 | 0.86 |
ENSDART00000114190
|
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr5_+_52039067 | 0.86 |
ENSDART00000143276
|
setbp1
|
SET binding protein 1 |
| chr3_-_37476475 | 0.85 |
ENSDART00000148107
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr5_-_38506981 | 0.85 |
ENSDART00000097822
|
atp1b2b
|
ATPase Na+/K+ transporting subunit beta 2b |
| chr13_-_32648382 | 0.85 |
ENSDART00000138571
ENSDART00000132820 |
bend3
|
BEN domain containing 3 |
| chr24_-_31123365 | 0.85 |
ENSDART00000182947
|
tmem56a
|
transmembrane protein 56a |
| chr5_-_65782783 | 0.85 |
ENSDART00000130888
ENSDART00000050855 |
notch1b
|
notch 1b |
| chr9_-_4598883 | 0.85 |
ENSDART00000171927
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr2_-_57076687 | 0.84 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr10_+_5159475 | 0.84 |
ENSDART00000142507
|
cdc42se2
|
CDC42 small effector 2 |
| chr8_-_22826996 | 0.84 |
ENSDART00000146563
|
magixa
|
MAGI family member, X-linked a |
| chr12_+_42574148 | 0.84 |
ENSDART00000157855
|
ebf3a
|
early B cell factor 3a |
| chr3_-_18373604 | 0.84 |
ENSDART00000148132
|
spag9a
|
sperm associated antigen 9a |
| chr13_+_12801092 | 0.83 |
ENSDART00000164040
ENSDART00000134725 |
si:ch211-233a24.2
|
si:ch211-233a24.2 |
| chr15_-_1962326 | 0.83 |
ENSDART00000179669
|
dock10
|
dedicator of cytokinesis 10 |
| chr5_-_18046053 | 0.82 |
ENSDART00000144898
|
rnf215
|
ring finger protein 215 |
| chr10_+_35953068 | 0.82 |
ENSDART00000015279
|
rtn4rl1a
|
reticulon 4 receptor-like 1a |
| chr18_-_22753637 | 0.82 |
ENSDART00000181589
ENSDART00000009912 |
hsf4
|
heat shock transcription factor 4 |
| chr9_-_11655031 | 0.82 |
ENSDART00000044314
|
itgav
|
integrin, alpha V |
| chr8_+_26565512 | 0.82 |
ENSDART00000140980
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr1_-_56080112 | 0.82 |
ENSDART00000075469
ENSDART00000161473 |
c3a.6
|
complement component c3a, duplicate 6 |
| chr1_-_46859398 | 0.82 |
ENSDART00000135795
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr25_-_13105310 | 0.81 |
ENSDART00000172269
|
smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr25_+_37126921 | 0.81 |
ENSDART00000124331
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr20_+_46897504 | 0.81 |
ENSDART00000158124
|
si:ch73-21k16.1
|
si:ch73-21k16.1 |
| chr18_-_14937211 | 0.81 |
ENSDART00000141893
|
mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr10_+_25222367 | 0.81 |
ENSDART00000042767
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr2_+_55916911 | 0.80 |
ENSDART00000189483
ENSDART00000183647 ENSDART00000083470 |
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr12_-_25217217 | 0.80 |
ENSDART00000152931
|
kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr10_+_439692 | 0.80 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr16_+_16849220 | 0.80 |
ENSDART00000047409
ENSDART00000142155 |
myh14
|
myosin, heavy chain 14, non-muscle |
| chr21_+_24536233 | 0.79 |
ENSDART00000145448
ENSDART00000109886 ENSDART00000135191 |
anapc13
|
anaphase promoting complex subunit 13 |
| chr15_-_36249087 | 0.79 |
ENSDART00000157190
|
ostn
|
osteocrin |
| chr3_+_16722014 | 0.79 |
ENSDART00000008711
|
gys1
|
glycogen synthase 1 (muscle) |
| chr5_-_41638039 | 0.79 |
ENSDART00000144525
|
epg5
|
ectopic P-granules autophagy protein 5 homolog (C. elegans) |
| chr19_-_3488860 | 0.78 |
ENSDART00000172520
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr3_-_2593859 | 0.78 |
ENSDART00000143826
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr7_+_34620418 | 0.78 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr1_-_21483832 | 0.78 |
ENSDART00000102790
|
glrba
|
glycine receptor, beta a |
| chr2_+_258698 | 0.77 |
ENSDART00000181330
ENSDART00000181645 |
phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr16_+_37470717 | 0.77 |
ENSDART00000112003
ENSDART00000188431 ENSDART00000192837 |
adgrb1a
|
adhesion G protein-coupled receptor B1a |
| chr2_-_14390627 | 0.77 |
ENSDART00000172367
|
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr9_-_27442339 | 0.76 |
ENSDART00000138602
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr14_+_8328645 | 0.76 |
ENSDART00000127494
|
nrg2b
|
neuregulin 2b |
| chr3_+_40841897 | 0.76 |
ENSDART00000150081
ENSDART00000126096 |
mmd2a
|
monocyte to macrophage differentiation-associated 2a |
| chr5_-_57641257 | 0.76 |
ENSDART00000149282
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr22_+_16446052 | 0.75 |
ENSDART00000142454
|
si:dkey-121a11.3
|
si:dkey-121a11.3 |
| chr7_-_30082931 | 0.75 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr8_+_25247245 | 0.75 |
ENSDART00000045798
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr9_+_40939336 | 0.74 |
ENSDART00000100386
|
mstnb
|
myostatin b |
| chr22_+_696931 | 0.74 |
ENSDART00000149712
ENSDART00000009756 |
gpr37l1a
|
G protein-coupled receptor 37 like 1a |
| chr23_+_21544227 | 0.74 |
ENSDART00000140253
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr20_+_38201644 | 0.73 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr12_+_2381213 | 0.73 |
ENSDART00000188007
|
LO018238.1
|
|
| chr22_+_5103349 | 0.73 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr19_+_27479838 | 0.73 |
ENSDART00000103922
|
atat1
|
alpha tubulin acetyltransferase 1 |
| chr17_+_16564921 | 0.73 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of klf12b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.5 | 1.6 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.5 | 2.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.4 | 2.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.3 | 1.2 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.3 | 2.0 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.3 | 1.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.3 | 0.8 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.3 | 0.8 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.3 | 2.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 0.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 1.4 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 3.6 | GO:0042026 | protein refolding(GO:0042026) |
| 0.2 | 2.6 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.2 | 0.9 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.2 | 0.6 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.2 | 1.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.8 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 0.7 | GO:1901862 | negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle tissue development(GO:1901862) |
| 0.2 | 0.9 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.2 | 0.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.7 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.2 | 1.9 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 0.7 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.2 | 0.5 | GO:2000793 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.2 | 1.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 1.5 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.1 | 0.4 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.1 | 1.5 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.1 | 0.9 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 1.9 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 2.8 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 0.4 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.9 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 1.0 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 1.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 1.7 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.7 | GO:0008584 | male gonad development(GO:0008584) |
| 0.1 | 0.4 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 0.3 | GO:2000376 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.1 | 1.5 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 0.9 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 1.0 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.1 | 0.9 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.1 | 0.3 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.8 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.8 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.5 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 0.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.8 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 1.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.8 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.0 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.5 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 1.1 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 1.3 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 0.5 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 0.8 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.5 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.8 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.9 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.1 | 1.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.9 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 1.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 3.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.4 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.5 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 1.7 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 1.3 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.3 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.3 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.2 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 2.5 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.8 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.9 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.3 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.5 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.8 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 1.4 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.6 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.3 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 2.1 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.3 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 2.0 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 3.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.6 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.5 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.0 | 5.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.6 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 1.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 1.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.7 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.2 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) mRNA methylation(GO:0080009) |
| 0.0 | 0.3 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 1.1 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 2.0 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 0.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.2 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.0 | 1.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.0 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 2.1 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.5 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.2 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 1.1 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.2 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.5 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.0 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.4 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.5 | GO:0034250 | positive regulation of cellular amide metabolic process(GO:0034250) positive regulation of translation(GO:0045727) |
| 0.0 | 0.8 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.8 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.5 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.5 | 1.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 2.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 1.7 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.2 | 1.2 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 2.2 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 1.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.9 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 1.0 | GO:0033270 | paranode region of axon(GO:0033270) juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 1.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 4.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 1.3 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 5.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 3.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 3.8 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 2.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.2 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.4 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.4 | 1.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.4 | 2.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.3 | 4.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 0.9 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.3 | 1.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 0.8 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.3 | 1.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 1.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 3.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 2.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 2.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 2.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 0.8 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 1.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 0.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 0.9 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 1.0 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.9 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 0.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 0.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.5 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.8 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 3.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.0 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 1.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.3 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.1 | 0.5 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 2.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.7 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.6 | GO:0031779 | melanocortin receptor binding(GO:0031779) |
| 0.1 | 1.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.8 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 2.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.2 | GO:0052833 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.8 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.3 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 0.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 5.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.3 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 0.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 2.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.7 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 1.0 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 1.1 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 1.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.7 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.4 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.9 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.8 | GO:0042626 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.1 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 1.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 3.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 1.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 1.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 2.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | ST GA13 PATHWAY | G alpha 13 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.1 | REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | Genes involved in Downstream signaling of activated FGFR |
| 0.4 | 1.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 2.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 1.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 2.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.0 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 0.8 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 3.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.8 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |