Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
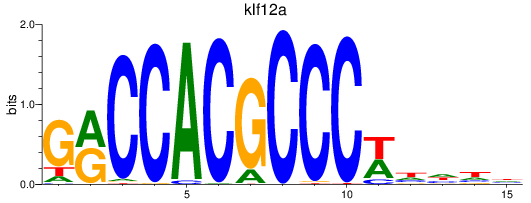
Results for klf12a
Z-value: 1.51
Transcription factors associated with klf12a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf12a
|
ENSDARG00000015312 | Kruppel-like factor 12a |
|
klf12a
|
ENSDARG00000115152 | Kruppel-like factor 12a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf12a | dr11_v1_chr1_+_34527213_34527213 | 0.45 | 5.1e-02 | Click! |
Activity profile of klf12a motif
Sorted Z-values of klf12a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_42132962 | 4.00 |
ENSDART00000187739
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr3_+_1211242 | 3.98 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr16_-_17197546 | 3.70 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr17_+_8925232 | 3.15 |
ENSDART00000036668
|
psmc1a
|
proteasome 26S subunit, ATPase 1a |
| chr13_+_28417297 | 3.02 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr4_+_17417111 | 2.88 |
ENSDART00000056005
|
ascl1a
|
achaete-scute family bHLH transcription factor 1a |
| chr25_+_3358701 | 2.82 |
ENSDART00000104877
|
chchd3b
|
coiled-coil-helix-coiled-coil-helix domain containing 3b |
| chr20_+_35058634 | 2.69 |
ENSDART00000122696
|
hnrnpub
|
heterogeneous nuclear ribonucleoprotein Ub |
| chr7_-_48665305 | 2.65 |
ENSDART00000190507
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr17_+_25332711 | 2.51 |
ENSDART00000082319
|
tmem54a
|
transmembrane protein 54a |
| chr17_-_14815557 | 2.47 |
ENSDART00000154473
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr13_-_33671694 | 2.42 |
ENSDART00000143945
ENSDART00000100504 |
zgc:163030
|
zgc:163030 |
| chr24_+_42127983 | 2.39 |
ENSDART00000190157
ENSDART00000176032 ENSDART00000175790 |
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr18_-_14677936 | 2.36 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr2_+_59015878 | 2.31 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr5_+_66132394 | 2.28 |
ENSDART00000073892
|
zgc:114041
|
zgc:114041 |
| chr3_-_50998577 | 2.20 |
ENSDART00000157735
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr3_-_32541033 | 2.19 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr21_-_308852 | 2.13 |
ENSDART00000151613
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr2_-_39675829 | 2.12 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr4_-_9852318 | 2.11 |
ENSDART00000080702
|
glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr18_-_49318823 | 2.10 |
ENSDART00000098419
|
sb:cb81
|
sb:cb81 |
| chr23_-_3759692 | 2.05 |
ENSDART00000028885
|
hmga1a
|
high mobility group AT-hook 1a |
| chr1_-_59232267 | 2.04 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr3_+_27027781 | 2.02 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr22_-_12626105 | 2.01 |
ENSDART00000115344
ENSDART00000193390 ENSDART00000183949 |
cep70
|
centrosomal protein 70 |
| chr23_-_31645760 | 1.97 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr22_+_5118361 | 1.95 |
ENSDART00000168371
ENSDART00000170222 ENSDART00000158846 |
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr7_+_38349667 | 1.95 |
ENSDART00000010046
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr14_+_33264303 | 1.89 |
ENSDART00000130680
ENSDART00000075187 |
pdzd11
|
PDZ domain containing 11 |
| chr9_-_1984604 | 1.85 |
ENSDART00000082339
|
hoxd12a
|
homeobox D12a |
| chr22_-_2922053 | 1.80 |
ENSDART00000178290
|
pak2b
|
p21 protein (Cdc42/Rac)-activated kinase 2b |
| chr11_-_25384213 | 1.77 |
ENSDART00000103650
|
mafbb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Bb |
| chr12_+_11080776 | 1.73 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr7_-_18601206 | 1.72 |
ENSDART00000111636
|
DTX4
|
si:ch211-119e14.2 |
| chr13_+_4405282 | 1.69 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr13_-_36621926 | 1.67 |
ENSDART00000057155
|
cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr10_+_15777064 | 1.66 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr22_-_775776 | 1.65 |
ENSDART00000149749
|
rbbp5
|
retinoblastoma binding protein 5 |
| chr8_-_4618653 | 1.64 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr22_-_21676364 | 1.64 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr2_-_27900518 | 1.64 |
ENSDART00000109561
ENSDART00000077720 |
zgc:163121
|
zgc:163121 |
| chr22_+_835728 | 1.62 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr25_+_3306858 | 1.60 |
ENSDART00000137077
|
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr13_-_6252498 | 1.60 |
ENSDART00000115157
|
tuba4l
|
tubulin, alpha 4 like |
| chr19_+_47394270 | 1.60 |
ENSDART00000171281
|
psmb2
|
proteasome subunit beta 2 |
| chr23_-_3759345 | 1.59 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr1_-_54107321 | 1.58 |
ENSDART00000148382
ENSDART00000150357 |
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr24_+_17005647 | 1.53 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr5_+_23136544 | 1.52 |
ENSDART00000003428
ENSDART00000109340 ENSDART00000171039 ENSDART00000178821 |
prps1a
|
phosphoribosyl pyrophosphate synthetase 1A |
| chr23_+_30730121 | 1.48 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr8_+_17167876 | 1.48 |
ENSDART00000134665
|
cenph
|
centromere protein H |
| chr19_+_29854223 | 1.45 |
ENSDART00000109729
ENSDART00000157419 |
si:ch73-130a3.4
|
si:ch73-130a3.4 |
| chr14_+_26439227 | 1.45 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr25_+_34013093 | 1.43 |
ENSDART00000011967
|
anxa2a
|
annexin A2a |
| chr6_+_16031189 | 1.42 |
ENSDART00000015333
|
gbx2
|
gastrulation brain homeobox 2 |
| chr11_+_29790626 | 1.41 |
ENSDART00000067822
|
dynlt3
|
dynein, light chain, Tctex-type 3 |
| chr22_-_18546241 | 1.39 |
ENSDART00000105404
ENSDART00000105405 |
cirbpb
|
cold inducible RNA binding protein b |
| chr14_+_24277556 | 1.38 |
ENSDART00000122660
|
hnrnpa0a
|
heterogeneous nuclear ribonucleoprotein A0a |
| chr18_+_41549764 | 1.38 |
ENSDART00000059135
|
bcl7bb
|
BCL tumor suppressor 7Bb |
| chr21_+_30549512 | 1.38 |
ENSDART00000132831
|
rab38c
|
RAB38c, member of RAS oncogene family |
| chr14_+_97017 | 1.38 |
ENSDART00000159300
ENSDART00000169523 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr22_-_881080 | 1.37 |
ENSDART00000185489
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr9_+_34425736 | 1.36 |
ENSDART00000135147
|
si:ch211-218d20.15
|
si:ch211-218d20.15 |
| chr23_-_6749820 | 1.35 |
ENSDART00000128772
|
ptges3b
|
prostaglandin E synthase 3b (cytosolic) |
| chr12_+_42436328 | 1.34 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr13_+_16279890 | 1.34 |
ENSDART00000101775
ENSDART00000057948 |
anxa11a
|
annexin A11a |
| chr18_-_46258612 | 1.33 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr8_+_20157798 | 1.32 |
ENSDART00000124809
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr3_+_23692462 | 1.30 |
ENSDART00000145934
|
hoxb7a
|
homeobox B7a |
| chr19_-_30420602 | 1.30 |
ENSDART00000144121
|
ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr12_+_48815988 | 1.29 |
ENSDART00000149089
|
anxa11b
|
annexin A11b |
| chr23_-_19831739 | 1.29 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr7_+_10563017 | 1.29 |
ENSDART00000193520
ENSDART00000173125 |
zfand6
|
zinc finger, AN1-type domain 6 |
| chr17_-_114121 | 1.28 |
ENSDART00000172408
ENSDART00000157784 |
arhgap11a
|
Rho GTPase activating protein 11A |
| chr23_-_27822920 | 1.28 |
ENSDART00000023094
|
acvr1ba
|
activin A receptor type 1Ba |
| chr18_+_44649804 | 1.27 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr7_-_59159253 | 1.26 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr2_-_32513538 | 1.26 |
ENSDART00000056640
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr19_-_42391383 | 1.26 |
ENSDART00000110075
ENSDART00000087002 |
plekho1a
|
pleckstrin homology domain containing, family O member 1a |
| chr25_+_3306620 | 1.25 |
ENSDART00000182085
ENSDART00000034704 |
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr24_-_21923930 | 1.25 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr19_+_46237665 | 1.24 |
ENSDART00000159391
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr17_-_14816465 | 1.23 |
ENSDART00000156326
ENSDART00000156287 |
nid2a
|
nidogen 2a (osteonidogen) |
| chr13_+_421231 | 1.22 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr14_-_6225336 | 1.21 |
ENSDART00000111681
|
hdx
|
highly divergent homeobox |
| chr8_+_17168114 | 1.21 |
ENSDART00000183901
|
cenph
|
centromere protein H |
| chr21_-_13856689 | 1.21 |
ENSDART00000102197
|
fam129ba
|
family with sequence similarity 129, member Ba |
| chr3_+_15893039 | 1.19 |
ENSDART00000055780
|
jpt2
|
Jupiter microtubule associated homolog 2 |
| chr16_-_21777545 | 1.19 |
ENSDART00000039381
|
cops7a
|
COP9 constitutive photomorphogenic homolog subunit 7A |
| chr13_+_18471546 | 1.18 |
ENSDART00000090712
ENSDART00000127962 |
stox1
|
storkhead box 1 |
| chr3_-_40528333 | 1.17 |
ENSDART00000193047
|
actb2
|
actin, beta 2 |
| chr17_+_16429826 | 1.17 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr22_-_4887401 | 1.16 |
ENSDART00000190841
|
si:ch73-256j6.4
|
si:ch73-256j6.4 |
| chr25_+_186583 | 1.15 |
ENSDART00000161504
|
pclaf
|
PCNA clamp associated factor |
| chr5_-_9824908 | 1.13 |
ENSDART00000169698
|
zgc:158343
|
zgc:158343 |
| chr15_-_5580093 | 1.13 |
ENSDART00000143726
|
wdr62
|
WD repeat domain 62 |
| chr20_-_25748407 | 1.12 |
ENSDART00000063152
|
chst14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 |
| chr5_-_36597612 | 1.11 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr15_+_47161917 | 1.11 |
ENSDART00000167860
|
gap43
|
growth associated protein 43 |
| chr25_+_19008497 | 1.11 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr16_-_45327616 | 1.11 |
ENSDART00000158733
|
si:dkey-33i11.1
|
si:dkey-33i11.1 |
| chr12_-_49168398 | 1.10 |
ENSDART00000186608
|
FO704624.1
|
|
| chr24_-_35534273 | 1.09 |
ENSDART00000026578
|
ube2v2
|
ubiquitin-conjugating enzyme E2 variant 2 |
| chr20_+_29565906 | 1.09 |
ENSDART00000062383
|
ywhaqa
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide a |
| chr4_-_11810365 | 1.08 |
ENSDART00000150529
|
cyb5r3
|
cytochrome b5 reductase 3 |
| chr10_+_8629275 | 1.08 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr16_+_28764017 | 1.08 |
ENSDART00000122433
|
trim33l
|
tripartite motif containing 33, like |
| chr17_+_44441042 | 1.07 |
ENSDART00000142123
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr5_+_26913120 | 1.06 |
ENSDART00000126609
|
tbx3b
|
T-box 3b |
| chr24_-_4450238 | 1.05 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr18_-_3166726 | 1.04 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr15_+_17752928 | 1.04 |
ENSDART00000155314
|
si:ch211-213d14.2
|
si:ch211-213d14.2 |
| chr12_-_48312647 | 1.03 |
ENSDART00000114415
|
ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr8_-_14179798 | 1.03 |
ENSDART00000040645
ENSDART00000146749 |
rhoaa
|
ras homolog gene family, member Aa |
| chr7_+_9880811 | 1.03 |
ENSDART00000128376
|
lins1
|
lines homolog 1 |
| chr6_-_18976168 | 1.03 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr2_+_30379650 | 1.01 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr17_-_24889837 | 1.01 |
ENSDART00000187980
|
gale
|
UDP-galactose-4-epimerase |
| chr5_-_72136548 | 1.00 |
ENSDART00000007827
|
spra
|
sepiapterin reductase a |
| chr16_+_26747766 | 1.00 |
ENSDART00000183257
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr10_+_15777258 | 1.00 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr11_-_36279602 | 1.00 |
ENSDART00000122531
ENSDART00000103064 ENSDART00000125616 |
nfya
|
nuclear transcription factor Y, alpha |
| chr1_-_59313465 | 0.99 |
ENSDART00000158067
ENSDART00000159419 |
txndc11
|
thioredoxin domain containing 11 |
| chr15_-_20468302 | 0.98 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr17_-_18898115 | 0.98 |
ENSDART00000028044
|
galcb
|
galactosylceramidase b |
| chr1_+_42874410 | 0.97 |
ENSDART00000153506
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr22_+_661711 | 0.97 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr19_-_34011340 | 0.97 |
ENSDART00000172618
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr4_+_9011448 | 0.97 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr19_-_35450661 | 0.96 |
ENSDART00000113574
ENSDART00000136895 |
anln
|
anillin, actin binding protein |
| chr7_-_20796935 | 0.96 |
ENSDART00000162357
|
dnajc3b
|
DnaJ (Hsp40) homolog, subfamily C, member 3b |
| chr6_-_25165693 | 0.95 |
ENSDART00000167259
|
znf326
|
zinc finger protein 326 |
| chr9_+_3035128 | 0.95 |
ENSDART00000140693
|
si:ch211-173m16.2
|
si:ch211-173m16.2 |
| chr3_+_32410746 | 0.95 |
ENSDART00000025496
|
rras
|
RAS related |
| chr17_-_20229812 | 0.94 |
ENSDART00000155989
|
ebf3b
|
early B cell factor 3b |
| chr6_+_12968101 | 0.94 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr19_-_31372896 | 0.94 |
ENSDART00000046609
|
scin
|
scinderin |
| chr23_+_44080382 | 0.93 |
ENSDART00000011715
|
zgc:56304
|
zgc:56304 |
| chr2_+_43894368 | 0.92 |
ENSDART00000143885
|
gbp3
|
guanylate binding protein 3 |
| chr14_+_80685 | 0.92 |
ENSDART00000188443
|
stag3
|
stromal antigen 3 |
| chr22_+_344763 | 0.92 |
ENSDART00000181934
|
CU914780.1
|
|
| chr4_-_4612116 | 0.92 |
ENSDART00000130601
|
CABZ01020840.1
|
Danio rerio apoptosis facilitator Bcl-2-like protein 14 (LOC101885512), mRNA. |
| chr17_-_31695217 | 0.91 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr5_+_36850650 | 0.90 |
ENSDART00000051186
|
nccrp1
|
non-specific cytotoxic cell receptor protein 1 |
| chr3_-_32362872 | 0.90 |
ENSDART00000035545
ENSDART00000012630 |
prmt1
|
protein arginine methyltransferase 1 |
| chr20_+_20672163 | 0.90 |
ENSDART00000027758
|
rtn1b
|
reticulon 1b |
| chr12_+_499881 | 0.90 |
ENSDART00000167527
|
mprip
|
myosin phosphatase Rho interacting protein |
| chr6_-_57938043 | 0.89 |
ENSDART00000171073
|
tox2
|
TOX high mobility group box family member 2 |
| chr4_-_11810799 | 0.89 |
ENSDART00000014153
|
cyb5r3
|
cytochrome b5 reductase 3 |
| chr3_-_31924643 | 0.87 |
ENSDART00000122589
|
rnf113a
|
ring finger protein 113A |
| chr14_+_28545198 | 0.87 |
ENSDART00000125362
ENSDART00000105902 |
hmmr
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr5_+_68807170 | 0.86 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr23_-_37085959 | 0.85 |
ENSDART00000139662
ENSDART00000031016 |
ints11
|
integrator complex subunit 11 |
| chr21_-_32060993 | 0.85 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr2_-_37837056 | 0.85 |
ENSDART00000158179
ENSDART00000160317 ENSDART00000171409 |
mettl17
|
methyltransferase like 17 |
| chr6_-_40657653 | 0.84 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr11_-_36341028 | 0.84 |
ENSDART00000146093
|
sort1a
|
sortilin 1a |
| chr23_+_10146542 | 0.83 |
ENSDART00000048073
|
zgc:171775
|
zgc:171775 |
| chr21_+_25236297 | 0.83 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr22_+_396840 | 0.83 |
ENSDART00000163198
|
capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr20_+_812012 | 0.82 |
ENSDART00000179299
ENSDART00000137135 |
mei4
|
meiosis-specific, MEI4 homolog (S. cerevisiae) |
| chr15_+_28410664 | 0.82 |
ENSDART00000132028
ENSDART00000057697 ENSDART00000057257 |
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr2_+_23222939 | 0.81 |
ENSDART00000026800
|
kifap3b
|
kinesin-associated protein 3b |
| chr12_+_6391243 | 0.80 |
ENSDART00000152765
|
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr9_+_23895711 | 0.80 |
ENSDART00000034686
|
cops8
|
COP9 signalosome subunit 8 |
| chr14_-_46442459 | 0.79 |
ENSDART00000158770
|
fgfbp1b
|
fibroblast growth factor binding protein 1b |
| chr1_+_45922699 | 0.79 |
ENSDART00000033669
|
lipt1
|
lipoyltransferase 1 |
| chr21_-_41369370 | 0.79 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr5_-_64431927 | 0.79 |
ENSDART00000158248
|
brd3b
|
bromodomain containing 3b |
| chr14_-_24277805 | 0.79 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr8_-_32385989 | 0.78 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr6_+_18520859 | 0.78 |
ENSDART00000158263
|
si:dkey-10p5.10
|
si:dkey-10p5.10 |
| chr20_-_46554440 | 0.77 |
ENSDART00000043298
ENSDART00000060680 |
fosab
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Ab |
| chr7_+_23907692 | 0.77 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr3_+_22984098 | 0.77 |
ENSDART00000043190
|
lsm12a
|
LSM12 homolog a |
| chr2_+_25658112 | 0.76 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr7_+_40083601 | 0.76 |
ENSDART00000099046
|
zgc:112356
|
zgc:112356 |
| chr19_+_27479838 | 0.76 |
ENSDART00000103922
|
atat1
|
alpha tubulin acetyltransferase 1 |
| chr3_+_726000 | 0.76 |
ENSDART00000158510
|
dicp1.17
|
diverse immunoglobulin domain-containing protein 1.17 |
| chr6_+_7533601 | 0.76 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr21_+_6197223 | 0.75 |
ENSDART00000147716
|
si:dkey-93m18.3
|
si:dkey-93m18.3 |
| chr1_-_54100988 | 0.75 |
ENSDART00000192662
|
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr10_-_8294965 | 0.75 |
ENSDART00000167380
|
plpp1a
|
phospholipid phosphatase 1a |
| chr17_-_30944566 | 0.74 |
ENSDART00000192663
|
evla
|
Enah/Vasp-like a |
| chr22_+_1947494 | 0.74 |
ENSDART00000159121
|
si:dkey-15h8.15
|
si:dkey-15h8.15 |
| chr14_-_1355544 | 0.74 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr9_+_1654284 | 0.73 |
ENSDART00000062854
|
nfe2l2a
|
nuclear factor, erythroid 2-like 2a |
| chr20_+_13894123 | 0.73 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr8_-_17167819 | 0.73 |
ENSDART00000135042
ENSDART00000143920 |
mrps36
|
mitochondrial ribosomal protein S36 |
| chr4_+_16710001 | 0.72 |
ENSDART00000035899
|
pkp2
|
plakophilin 2 |
| chr2_+_20967673 | 0.72 |
ENSDART00000057174
|
arpc5a
|
actin related protein 2/3 complex, subunit 5A |
| chr11_-_11301283 | 0.71 |
ENSDART00000113311
ENSDART00000180466 |
col9a1a
|
collagen, type IX, alpha 1a |
| chr7_-_48396193 | 0.71 |
ENSDART00000083555
|
sin3ab
|
SIN3 transcription regulator family member Ab |
| chr2_+_25657958 | 0.70 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr24_-_11076400 | 0.70 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr9_+_30421489 | 0.70 |
ENSDART00000145025
ENSDART00000132058 |
zgc:113314
|
zgc:113314 |
| chr16_-_7793457 | 0.69 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr13_-_44808783 | 0.69 |
ENSDART00000099984
|
glo1
|
glyoxalase 1 |
| chr7_-_31938938 | 0.68 |
ENSDART00000132353
|
bdnf
|
brain-derived neurotrophic factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of klf12a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.7 | 2.9 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.7 | 2.0 | GO:0003093 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.7 | 2.7 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.7 | 2.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.4 | 1.5 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.4 | 2.6 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.4 | 1.9 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.4 | 1.1 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.3 | 1.1 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.2 | 0.7 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.2 | 1.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 1.0 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.2 | 2.1 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.2 | 0.9 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.2 | 0.7 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 2.2 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 2.3 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 | 1.0 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.2 | 1.1 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.2 | 1.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 1.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.2 | 2.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 0.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 0.8 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.2 | 1.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.0 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 1.4 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.1 | 1.0 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.7 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 1.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 1.9 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 1.3 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 1.0 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.7 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.1 | 1.4 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 1.6 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.1 | 2.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.8 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 2.8 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.1 | 1.6 | GO:0042559 | pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.1 | 0.9 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.8 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.7 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 3.7 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 0.6 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 2.6 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.1 | 0.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 2.2 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 0.9 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.1 | 1.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 3.7 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.9 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) cellular response to fatty acid(GO:0071398) |
| 0.1 | 0.4 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 0.6 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.4 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.1 | 0.6 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.1 | 0.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.5 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.3 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.4 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.1 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 1.0 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.5 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 1.4 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 1.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 1.5 | GO:0072662 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 6.4 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.1 | 1.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 1.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.6 | GO:0033260 | nuclear DNA replication(GO:0033260) |
| 0.1 | 0.6 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 0.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.7 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.7 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 1.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.0 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.6 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.6 | GO:0051963 | regulation of synapse assembly(GO:0051963) |
| 0.0 | 0.2 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 0.9 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.9 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.4 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 1.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 2.7 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 2.6 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.7 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.6 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.9 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 1.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.3 | GO:0061098 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of protein tyrosine kinase activity(GO:0061098) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.8 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.3 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.4 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 1.3 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.0 | 1.6 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.6 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.1 | GO:0035143 | caudal fin morphogenesis(GO:0035143) |
| 0.0 | 1.1 | GO:0003205 | cardiac chamber development(GO:0003205) |
| 0.0 | 3.5 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.5 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 2.0 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.3 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.6 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.7 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 1.0 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.8 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.9 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 3.7 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 0.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 1.0 | GO:0006476 | protein deacetylation(GO:0006476) |
| 0.0 | 0.4 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.2 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 1.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.2 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 1.4 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.5 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.6 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.4 | 1.5 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.3 | 2.6 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.3 | 1.6 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 1.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.3 | 2.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 0.7 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 1.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 3.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.2 | 1.5 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.2 | 1.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 2.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 0.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 2.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.5 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.1 | 1.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 2.7 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 5.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 4.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.9 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 2.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.9 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 1.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 3.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.5 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.3 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.5 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 4.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 2.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.7 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.6 | 1.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.5 | 2.7 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.5 | 3.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.5 | 2.3 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.4 | 1.2 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.4 | 1.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.3 | 1.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.3 | 1.0 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.3 | 1.0 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.3 | 2.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 1.6 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.3 | 2.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 1.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.3 | 2.0 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.3 | 2.0 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.3 | 1.4 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 1.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.2 | 1.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 0.6 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.2 | 2.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.2 | 1.0 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.8 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 2.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.5 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 1.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.1 | 1.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.3 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 1.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.7 | GO:0008200 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.1 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.8 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.6 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.4 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.7 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.9 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 0.7 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 1.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 2.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.5 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 1.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 2.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.6 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 1.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 1.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.4 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 9.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 7.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.3 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.8 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 1.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 3.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 1.1 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.0 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.6 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 5.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 3.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 0.9 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 2.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 7.2 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.1 | 0.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 2.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 1.6 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 1.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.1 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |