Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for jund_batf
Z-value: 0.80
Transcription factors associated with jund_batf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
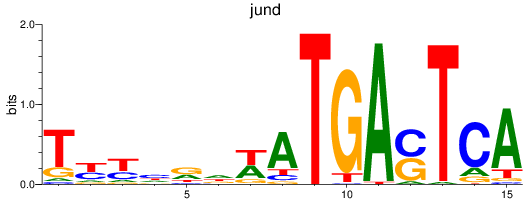
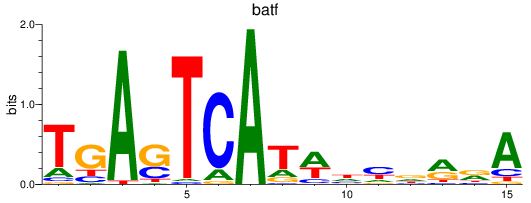
jund
|
ENSDARG00000067850 | JunD proto-oncogene, AP-1 transcription factor subunit |
|
batf
|
ENSDARG00000011818 | basic leucine zipper transcription factor, ATF-like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| batf | dr11_v1_chr20_+_46572550_46572550 | -0.33 | 1.7e-01 | Click! |
| jund | dr11_v1_chr2_-_56131312_56131312 | -0.31 | 1.9e-01 | Click! |
Activity profile of jund_batf motif
Sorted Z-values of jund_batf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_42972599 | 2.06 |
ENSDART00000100751
|
pomcb
|
proopiomelanocortin b |
| chr3_-_26017592 | 1.81 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr3_-_26017831 | 1.60 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr16_-_24832038 | 1.36 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr7_-_26436436 | 1.31 |
ENSDART00000019035
ENSDART00000123395 |
her8a
|
hairy-related 8a |
| chr22_+_16497670 | 1.27 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr23_-_1017428 | 1.24 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr7_-_52963493 | 1.21 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr18_-_30499489 | 1.17 |
ENSDART00000033746
|
gins2
|
GINS complex subunit 2 |
| chr24_+_31361407 | 1.17 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr4_+_23127284 | 1.07 |
ENSDART00000122675
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr13_+_35637875 | 0.97 |
ENSDART00000180657
|
thbs2a
|
thrombospondin 2a |
| chr23_+_30967686 | 0.97 |
ENSDART00000144485
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr10_+_28428222 | 0.92 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr4_+_68087932 | 0.92 |
ENSDART00000150494
|
znf1096
|
zinc finger protein 1096 |
| chr13_-_50624743 | 0.92 |
ENSDART00000167949
|
vox
|
ventral homeobox |
| chr21_-_44512893 | 0.89 |
ENSDART00000166853
|
zgc:136410
|
zgc:136410 |
| chr3_+_31177972 | 0.86 |
ENSDART00000185954
|
clec19a
|
C-type lectin domain containing 19A |
| chr22_-_26595027 | 0.85 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr23_-_1017605 | 0.81 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr3_-_16289826 | 0.79 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr11_-_39202915 | 0.78 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr25_-_29072162 | 0.77 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr4_+_23127104 | 0.76 |
ENSDART00000139543
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr24_+_17260329 | 0.74 |
ENSDART00000129554
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr19_-_5669122 | 0.73 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr25_-_11026907 | 0.72 |
ENSDART00000156846
|
mespbb
|
mesoderm posterior bb |
| chr24_+_35827766 | 0.70 |
ENSDART00000144700
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr3_-_30488063 | 0.70 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr9_+_38644976 | 0.70 |
ENSDART00000133849
ENSDART00000135774 |
slc12a8
|
solute carrier family 12, member 8 |
| chr7_+_6317866 | 0.69 |
ENSDART00000173397
|
si:ch211-220f21.3
|
si:ch211-220f21.3 |
| chr20_+_572037 | 0.69 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr24_+_17260001 | 0.66 |
ENSDART00000066765
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr20_-_30931139 | 0.66 |
ENSDART00000006778
ENSDART00000146376 |
acat2
|
acetyl-CoA acetyltransferase 2 |
| chr2_+_52049239 | 0.64 |
ENSDART00000036813
|
ccdc94
|
coiled-coil domain containing 94 |
| chr8_-_52745141 | 0.64 |
ENSDART00000168359
ENSDART00000168252 |
fgf17
|
fibroblast growth factor 17 |
| chr7_-_13906409 | 0.62 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr11_+_3499111 | 0.62 |
ENSDART00000113096
ENSDART00000171621 |
pusl1
|
pseudouridylate synthase-like 1 |
| chr13_-_50624173 | 0.60 |
ENSDART00000184181
|
vox
|
ventral homeobox |
| chr4_+_1722899 | 0.58 |
ENSDART00000021139
|
fgfr1op2
|
FGFR1 oncogene partner 2 |
| chr8_-_36554675 | 0.57 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr17_-_6536466 | 0.57 |
ENSDART00000188735
|
cenpo
|
centromere protein O |
| chr11_+_7276983 | 0.56 |
ENSDART00000172407
ENSDART00000161176 ENSDART00000161041 ENSDART00000165536 |
si:ch73-238c9.1
|
si:ch73-238c9.1 |
| chr21_+_13233377 | 0.55 |
ENSDART00000142569
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr20_+_34868933 | 0.54 |
ENSDART00000153006
|
ankef1a
|
ankyrin repeat and EF-hand domain containing 1a |
| chr8_+_26432677 | 0.51 |
ENSDART00000078369
ENSDART00000131925 |
zgc:136971
|
zgc:136971 |
| chr18_+_9362455 | 0.51 |
ENSDART00000187025
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr14_-_22100118 | 0.51 |
ENSDART00000157547
|
ssrp1a
|
structure specific recognition protein 1a |
| chr15_-_18115540 | 0.51 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr8_-_38201415 | 0.50 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr16_-_25608453 | 0.49 |
ENSDART00000140140
|
zgc:110410
|
zgc:110410 |
| chr19_+_37118547 | 0.48 |
ENSDART00000103163
|
cx30.9
|
connexin 30.9 |
| chr5_+_50953240 | 0.47 |
ENSDART00000148501
ENSDART00000149892 ENSDART00000190312 |
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr18_-_46010 | 0.46 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr3_+_31621774 | 0.46 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr4_+_54899568 | 0.45 |
ENSDART00000162786
|
si:dkey-56m15.8
|
si:dkey-56m15.8 |
| chr8_+_29856013 | 0.45 |
ENSDART00000061981
ENSDART00000149610 |
hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr1_-_56176976 | 0.45 |
ENSDART00000052688
|
c3a.1
|
complement component c3a, duplicate 1 |
| chr13_-_5257303 | 0.45 |
ENSDART00000110610
|
si:dkey-78p8.1
|
si:dkey-78p8.1 |
| chr5_+_13385837 | 0.44 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr9_+_33334501 | 0.43 |
ENSDART00000006867
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr12_-_21684197 | 0.42 |
ENSDART00000152999
ENSDART00000153109 ENSDART00000148698 |
eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr3_-_41535647 | 0.40 |
ENSDART00000153723
ENSDART00000154198 |
si:ch211-222n22.1
|
si:ch211-222n22.1 |
| chr12_+_19188542 | 0.39 |
ENSDART00000134726
ENSDART00000148011 ENSDART00000109541 |
cby1
|
chibby homolog 1 (Drosophila) |
| chr12_+_1455147 | 0.39 |
ENSDART00000018752
|
cops3
|
COP9 signalosome subunit 3 |
| chr4_-_45146463 | 0.39 |
ENSDART00000187483
|
znf1046
|
zinc finger protein 1046 |
| chr19_+_48102560 | 0.38 |
ENSDART00000164464
|
utp18
|
UTP18 small subunit (SSU) processome component |
| chr2_-_24069331 | 0.38 |
ENSDART00000156972
ENSDART00000181691 ENSDART00000157041 |
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr16_+_37876779 | 0.37 |
ENSDART00000140148
|
si:ch211-198c19.1
|
si:ch211-198c19.1 |
| chr14_-_16807206 | 0.36 |
ENSDART00000157957
|
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr8_+_39634114 | 0.35 |
ENSDART00000144293
|
msi1
|
musashi RNA-binding protein 1 |
| chr24_-_24797455 | 0.35 |
ENSDART00000138741
|
pde7a
|
phosphodiesterase 7A |
| chr7_+_25036188 | 0.34 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr20_-_15922210 | 0.34 |
ENSDART00000152412
ENSDART00000152354 ENSDART00000152828 ENSDART00000013453 ENSDART00000152357 |
fam20b
|
family with sequence similarity 20, member B (H. sapiens) |
| chr13_+_1872767 | 0.33 |
ENSDART00000161162
|
bmp5
|
bone morphogenetic protein 5 |
| chr25_+_18583877 | 0.33 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr16_+_46410520 | 0.33 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr15_+_44283723 | 0.32 |
ENSDART00000167722
|
cwf19l2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr10_+_7718156 | 0.31 |
ENSDART00000189101
|
ggcx
|
gamma-glutamyl carboxylase |
| chr5_+_24089334 | 0.31 |
ENSDART00000183748
|
tp53
|
tumor protein p53 |
| chr12_+_41348969 | 0.30 |
ENSDART00000171352
|
si:ch211-27e6.1
|
si:ch211-27e6.1 |
| chr15_-_31357634 | 0.30 |
ENSDART00000127485
|
or111-2
|
odorant receptor, family D, subfamily 111, member 2 |
| chr25_-_24233884 | 0.30 |
ENSDART00000146419
|
si:dkey-11e23.9
|
si:dkey-11e23.9 |
| chr20_+_25340814 | 0.29 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr17_-_6536305 | 0.29 |
ENSDART00000154855
|
cenpo
|
centromere protein O |
| chr19_+_26072624 | 0.29 |
ENSDART00000147627
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr20_-_34768301 | 0.29 |
ENSDART00000142482
|
paqr8
|
progestin and adipoQ receptor family member VIII |
| chr9_-_1978090 | 0.29 |
ENSDART00000082344
|
hoxd11a
|
homeobox D11a |
| chr4_-_50434519 | 0.29 |
ENSDART00000150372
|
znf1061
|
zinc finger protein 1061 |
| chr6_+_7421898 | 0.29 |
ENSDART00000043946
|
ccdc65
|
coiled-coil domain containing 65 |
| chr10_-_40514643 | 0.29 |
ENSDART00000140705
|
taar19k
|
trace amine associated receptor 19k |
| chr23_+_19655301 | 0.28 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr12_+_21684239 | 0.28 |
ENSDART00000043546
|
mrpl27
|
mitochondrial ribosomal protein L27 |
| chr13_-_42749916 | 0.28 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr7_-_39751540 | 0.28 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr23_+_10431776 | 0.27 |
ENSDART00000144267
|
eif4ba
|
eukaryotic translation initiation factor 4Ba |
| chr3_+_60044780 | 0.27 |
ENSDART00000080437
|
zgc:113030
|
zgc:113030 |
| chr13_-_12602920 | 0.27 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr21_+_25765734 | 0.27 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr22_+_19218733 | 0.27 |
ENSDART00000183212
ENSDART00000133595 |
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr13_-_43108693 | 0.27 |
ENSDART00000164439
|
si:ch211-106f21.1
|
si:ch211-106f21.1 |
| chr4_+_68088529 | 0.26 |
ENSDART00000159155
|
znf1096
|
zinc finger protein 1096 |
| chr12_+_9551667 | 0.26 |
ENSDART00000048493
ENSDART00000166178 |
p4ha1a
|
prolyl 4-hydroxylase, alpha polypeptide I a |
| chr17_-_33714636 | 0.26 |
ENSDART00000188500
|
DNAL1
|
si:dkey-84k17.3 |
| chr25_-_18330503 | 0.26 |
ENSDART00000104496
|
dusp6
|
dual specificity phosphatase 6 |
| chr15_-_21877726 | 0.25 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr14_+_22132896 | 0.25 |
ENSDART00000138274
|
ccng1
|
cyclin G1 |
| chr11_+_7276824 | 0.25 |
ENSDART00000185992
|
si:ch73-238c9.1
|
si:ch73-238c9.1 |
| chr23_-_44848961 | 0.25 |
ENSDART00000136839
|
wu:fb72h05
|
wu:fb72h05 |
| chr23_-_4975452 | 0.24 |
ENSDART00000105241
ENSDART00000169978 |
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr7_-_24236364 | 0.24 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr4_+_42175261 | 0.24 |
ENSDART00000162193
|
si:ch211-142b24.2
|
si:ch211-142b24.2 |
| chr23_+_29883216 | 0.24 |
ENSDART00000113367
ENSDART00000149761 ENSDART00000148706 |
aurkaip1
|
aurora kinase A interacting protein 1 |
| chr4_+_28356606 | 0.23 |
ENSDART00000192995
|
si:ch73-263o4.4
|
si:ch73-263o4.4 |
| chr25_+_29474583 | 0.23 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr5_+_66170479 | 0.23 |
ENSDART00000172117
|
gldc
|
glycine dehydrogenase (decarboxylating) |
| chr5_-_30924347 | 0.23 |
ENSDART00000111749
ENSDART00000086564 ENSDART00000153909 |
spns2
|
spinster homolog 2 (Drosophila) |
| chr25_+_27873671 | 0.22 |
ENSDART00000088817
|
iqub
|
IQ motif and ubiquitin domain containing |
| chr4_+_64549970 | 0.22 |
ENSDART00000167846
|
si:ch211-223a21.3
|
si:ch211-223a21.3 |
| chr2_+_27855102 | 0.22 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr9_-_23181062 | 0.22 |
ENSDART00000192578
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr16_+_19029297 | 0.22 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr6_+_59832786 | 0.22 |
ENSDART00000154985
ENSDART00000102148 |
ddx3b
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3b |
| chr3_-_16493528 | 0.22 |
ENSDART00000142099
|
si:ch211-23l10.3
|
si:ch211-23l10.3 |
| chr13_+_10621257 | 0.21 |
ENSDART00000008603
|
prepl
|
prolyl endopeptidase-like |
| chr7_-_35408618 | 0.21 |
ENSDART00000074963
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr14_+_48062180 | 0.21 |
ENSDART00000056713
|
ppid
|
peptidylprolyl isomerase D |
| chr7_+_55149001 | 0.21 |
ENSDART00000148642
|
cdh31
|
cadherin 31 |
| chr23_+_29570386 | 0.21 |
ENSDART00000193192
|
lzic
|
leucine zipper and CTNNBIP1 domain containing |
| chr22_-_817479 | 0.20 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr10_+_36695597 | 0.20 |
ENSDART00000169015
ENSDART00000171392 |
rab6a
|
RAB6A, member RAS oncogene family |
| chr21_+_8341774 | 0.20 |
ENSDART00000129749
ENSDART00000055325 ENSDART00000133804 |
psmb7
|
proteasome subunit beta 7 |
| chr5_-_43959972 | 0.19 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr6_-_31224563 | 0.19 |
ENSDART00000104616
|
lepr
|
leptin receptor |
| chr8_+_25900049 | 0.19 |
ENSDART00000124300
ENSDART00000127618 ENSDART00000024009 |
rhoab
|
ras homolog gene family, member Ab |
| chr14_-_17588345 | 0.18 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr6_+_49095646 | 0.18 |
ENSDART00000103385
|
slc25a55a
|
solute carrier family 25, member 55a |
| chr20_+_20726231 | 0.18 |
ENSDART00000147112
|
zgc:193541
|
zgc:193541 |
| chr2_+_36608387 | 0.18 |
ENSDART00000159541
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr3_+_30968176 | 0.18 |
ENSDART00000186266
|
prf1.9
|
perforin 1.9 |
| chr15_+_29123031 | 0.17 |
ENSDART00000133988
ENSDART00000060030 |
zgc:101731
|
zgc:101731 |
| chr2_+_25378457 | 0.17 |
ENSDART00000089108
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr21_+_25071805 | 0.17 |
ENSDART00000078651
|
dixdc1b
|
DIX domain containing 1b |
| chr3_-_39488639 | 0.17 |
ENSDART00000161644
|
zgc:100868
|
zgc:100868 |
| chr16_-_5105295 | 0.17 |
ENSDART00000082071
ENSDART00000148955 ENSDART00000184700 ENSDART00000188127 |
bckdhb
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr25_+_27873836 | 0.17 |
ENSDART00000163801
|
iqub
|
IQ motif and ubiquitin domain containing |
| chr6_-_48317562 | 0.17 |
ENSDART00000103425
|
st7l
|
suppression of tumorigenicity 7 like |
| chr23_-_36753195 | 0.16 |
ENSDART00000181104
|
CU571398.1
|
|
| chr18_-_36909773 | 0.16 |
ENSDART00000141694
|
si:ch211-160d20.5
|
si:ch211-160d20.5 |
| chr8_-_26033176 | 0.16 |
ENSDART00000184533
|
BX784025.1
|
|
| chr17_+_33495194 | 0.15 |
ENSDART00000033691
|
pth2
|
parathyroid hormone 2 |
| chr5_-_41531629 | 0.15 |
ENSDART00000051082
|
akr1a1a
|
aldo-keto reductase family 1, member A1a (aldehyde reductase) |
| chr18_-_45761868 | 0.15 |
ENSDART00000025423
|
cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr20_-_2134620 | 0.15 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr22_+_19266995 | 0.15 |
ENSDART00000133995
ENSDART00000144963 |
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr2_+_47471647 | 0.15 |
ENSDART00000184199
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr8_+_8973425 | 0.15 |
ENSDART00000066107
|
bcap31
|
B cell receptor associated protein 31 |
| chr8_-_7603700 | 0.15 |
ENSDART00000137975
|
irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr7_+_69449814 | 0.15 |
ENSDART00000109644
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr25_+_10410620 | 0.14 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr3_-_21402279 | 0.14 |
ENSDART00000164513
|
CT573446.1
|
|
| chr5_-_24270989 | 0.14 |
ENSDART00000146251
|
si:ch211-137i24.12
|
si:ch211-137i24.12 |
| chr3_+_53352018 | 0.14 |
ENSDART00000082715
|
camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr18_+_45114392 | 0.14 |
ENSDART00000172328
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr4_+_13586689 | 0.14 |
ENSDART00000067161
ENSDART00000138201 |
tnpo3
|
transportin 3 |
| chr16_-_47427016 | 0.14 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr10_+_42733210 | 0.14 |
ENSDART00000189832
|
CABZ01063556.1
|
|
| chr16_+_42830152 | 0.14 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr11_+_45219558 | 0.13 |
ENSDART00000167828
|
tmc6b
|
transmembrane channel-like 6b |
| chr17_+_33496067 | 0.13 |
ENSDART00000184724
|
pth2
|
parathyroid hormone 2 |
| chr12_+_28117365 | 0.13 |
ENSDART00000066290
|
UTS2R
|
urotensin 2 receptor |
| chr1_-_53407448 | 0.13 |
ENSDART00000160033
ENSDART00000172322 |
elmod2
|
ELMO/CED-12 domain containing 2 |
| chr12_-_7639120 | 0.13 |
ENSDART00000126712
ENSDART00000126219 |
ccdc6b
|
coiled-coil domain containing 6b |
| chr16_-_47426482 | 0.13 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr1_+_54835131 | 0.13 |
ENSDART00000145070
|
si:ch211-196h16.4
|
si:ch211-196h16.4 |
| chr3_+_58092212 | 0.12 |
ENSDART00000156059
|
si:ch211-256e16.7
|
si:ch211-256e16.7 |
| chr20_+_53474963 | 0.12 |
ENSDART00000138976
|
bub1ba
|
BUB1 mitotic checkpoint serine/threonine kinase Ba |
| chr7_-_22699716 | 0.12 |
ENSDART00000193773
|
BX470211.5
|
|
| chr6_+_56141852 | 0.12 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr10_+_31953502 | 0.12 |
ENSDART00000185634
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr3_-_39488482 | 0.12 |
ENSDART00000135192
|
zgc:100868
|
zgc:100868 |
| chr4_-_64605318 | 0.12 |
ENSDART00000170391
|
znf1099
|
zinc finger protein 1099 |
| chr25_+_29474982 | 0.12 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr23_-_10914275 | 0.12 |
ENSDART00000112965
|
pdzrn3a
|
PDZ domain containing RING finger 3a |
| chr9_+_38372216 | 0.12 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr19_+_7552699 | 0.12 |
ENSDART00000180788
ENSDART00000115058 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr22_-_12746539 | 0.12 |
ENSDART00000175374
|
plcd4a
|
phospholipase C, delta 4a |
| chr11_-_18020258 | 0.11 |
ENSDART00000156116
|
qrich1
|
glutamine-rich 1 |
| chr13_+_43639867 | 0.11 |
ENSDART00000042588
ENSDART00000074728 |
zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr19_+_10527228 | 0.11 |
ENSDART00000091918
|
si:ch73-160i9.3
|
si:ch73-160i9.3 |
| chr9_-_24244383 | 0.10 |
ENSDART00000182407
|
cavin2a
|
caveolae associated protein 2a |
| chr25_-_19486399 | 0.10 |
ENSDART00000155076
ENSDART00000156016 |
zgc:193812
|
zgc:193812 |
| chr10_-_40479911 | 0.10 |
ENSDART00000136741
|
taar20d1
|
trace amine associated receptor 20d1 |
| chr25_+_22281441 | 0.10 |
ENSDART00000089516
|
stoml1
|
stomatin (EPB72)-like 1 |
| chr25_-_11378623 | 0.10 |
ENSDART00000166586
|
enc2
|
ectodermal-neural cortex 2 |
| chr3_-_4760384 | 0.10 |
ENSDART00000108810
|
CABZ01046997.1
|
|
| chr7_-_35409027 | 0.09 |
ENSDART00000128334
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr14_-_493029 | 0.09 |
ENSDART00000115093
|
zgc:172215
|
zgc:172215 |
| chr10_+_36328702 | 0.09 |
ENSDART00000166676
|
or106-2
|
odorant receptor, family G, subfamily 106, member 2 |
| chr8_+_25893071 | 0.09 |
ENSDART00000078161
|
tmem115
|
transmembrane protein 115 |
Network of associatons between targets according to the STRING database.
First level regulatory network of jund_batf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.4 | 1.8 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.2 | 0.5 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.2 | 0.5 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.2 | 0.5 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.2 | 1.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.5 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 1.5 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.7 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.3 | GO:0010525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.6 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 1.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.7 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.5 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 1.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.6 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) negative regulation of endodermal cell fate specification(GO:0042664) |
| 0.1 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.3 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.3 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 | 2.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.4 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.5 | GO:0046661 | male sex differentiation(GO:0046661) |
| 0.0 | 0.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.7 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.2 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.3 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.0 | 0.3 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.4 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 1.0 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.1 | GO:0021519 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0010935 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of response to cytokine stimulus(GO:0060760) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.7 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 1.4 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.1 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.1 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.1 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.2 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 1.2 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) cellular response to interleukin-1(GO:0071347) |
| 0.0 | 0.1 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 0.9 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.1 | 1.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.2 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 1.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.2 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.5 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.2 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 0.7 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 3.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.3 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 1.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.3 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.8 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 2.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.5 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.3 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 1.5 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 0.6 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 1.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |