Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
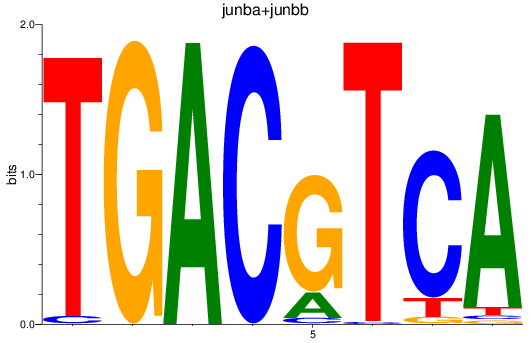
Results for junba+junbb
Z-value: 2.10
Transcription factors associated with junba+junbb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
junba
|
ENSDARG00000074378 | JunB proto-oncogene, AP-1 transcription factor subunit a |
|
junbb
|
ENSDARG00000104773 | JunB proto-oncogene, AP-1 transcription factor subunit b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| junba | dr11_v1_chr1_-_51734524_51734535 | 0.71 | 7.3e-04 | Click! |
| junbb | dr11_v1_chr3_-_7656059_7656059 | 0.42 | 7.1e-02 | Click! |
Activity profile of junba+junbb motif
Sorted Z-values of junba+junbb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_12385308 | 9.70 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr7_+_39385540 | 6.66 |
ENSDART00000139240
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr15_-_34418525 | 6.39 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr5_-_26118855 | 6.34 |
ENSDART00000009028
|
ela3l
|
elastase 3 like |
| chr4_-_16853464 | 6.13 |
ENSDART00000125743
ENSDART00000164570 |
slc25a3a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3a |
| chr2_-_43168292 | 5.36 |
ENSDART00000132588
|
crema
|
cAMP responsive element modulator a |
| chr15_+_45643787 | 5.32 |
ENSDART00000055995
ENSDART00000157750 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr15_-_12319065 | 5.25 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr16_-_383664 | 5.18 |
ENSDART00000051693
|
irx4a
|
iroquois homeobox 4a |
| chr23_+_19590006 | 5.15 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr6_+_27667359 | 4.91 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr13_+_23988442 | 4.90 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr2_-_11512819 | 4.89 |
ENSDART00000142013
|
penka
|
proenkephalin a |
| chr12_-_17707449 | 4.88 |
ENSDART00000142427
ENSDART00000034914 |
pvalb3
|
parvalbumin 3 |
| chr16_-_12319822 | 4.83 |
ENSDART00000127453
ENSDART00000184526 |
trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr19_-_10554240 | 4.63 |
ENSDART00000104539
ENSDART00000144923 |
lim2.4
|
lens intrinsic membrane protein 2.4 |
| chr12_+_5129245 | 4.52 |
ENSDART00000169073
|
pde6c
|
phosphodiesterase 6C, cGMP-specific, cone, alpha prime |
| chr15_-_44512461 | 4.50 |
ENSDART00000155456
|
gria4a
|
glutamate receptor, ionotropic, AMPA 4a |
| chr9_+_22003942 | 4.43 |
ENSDART00000091013
|
si:dkey-57a22.15
|
si:dkey-57a22.15 |
| chr25_+_16945348 | 4.42 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr23_+_19590598 | 4.41 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr16_-_43026273 | 4.07 |
ENSDART00000156820
ENSDART00000189080 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr10_+_17026870 | 4.05 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr21_-_43606502 | 4.02 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr16_-_17207754 | 4.01 |
ENSDART00000063804
|
wu:fj39g12
|
wu:fj39g12 |
| chr7_-_26457208 | 3.95 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr1_-_54947592 | 3.79 |
ENSDART00000129710
|
crtac1a
|
cartilage acidic protein 1a |
| chr14_+_11430796 | 3.77 |
ENSDART00000165275
|
si:ch211-153b23.3
|
si:ch211-153b23.3 |
| chr3_-_13147310 | 3.77 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr19_-_6988837 | 3.76 |
ENSDART00000145741
ENSDART00000167640 |
znf384l
|
zinc finger protein 384 like |
| chr3_-_36115339 | 3.70 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr7_+_69841017 | 3.61 |
ENSDART00000169107
|
FO818704.1
|
|
| chr18_+_910992 | 3.55 |
ENSDART00000161206
ENSDART00000167229 |
pkma
|
pyruvate kinase M1/2a |
| chr9_+_219124 | 3.50 |
ENSDART00000161484
|
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr3_+_32403758 | 3.49 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr19_-_10196370 | 3.47 |
ENSDART00000091707
|
dbpa
|
D site albumin promoter binding protein a |
| chr9_+_42095220 | 3.45 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr7_+_10592152 | 3.37 |
ENSDART00000182624
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr5_-_24238733 | 3.31 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr18_+_21408794 | 3.31 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr8_+_23174137 | 3.28 |
ENSDART00000189470
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr19_-_13808630 | 3.25 |
ENSDART00000166895
ENSDART00000187670 |
ctgfb
|
connective tissue growth factor b |
| chr16_-_43025885 | 3.21 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr23_+_45584223 | 3.17 |
ENSDART00000149367
|
si:ch73-290k24.5
|
si:ch73-290k24.5 |
| chr23_+_17220986 | 3.16 |
ENSDART00000054761
|
nol4lb
|
nucleolar protein 4-like b |
| chr8_+_49975160 | 3.15 |
ENSDART00000156403
ENSDART00000080135 |
gfpt1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr22_+_28337429 | 3.15 |
ENSDART00000166177
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr17_-_47090440 | 3.15 |
ENSDART00000163542
|
CABZ01056321.1
|
|
| chr15_-_163586 | 3.10 |
ENSDART00000163597
|
SEPT4
|
septin-4 |
| chr18_-_38087875 | 3.10 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr20_-_36671660 | 3.08 |
ENSDART00000134819
|
slc5a6a
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6a |
| chr24_-_7632187 | 3.06 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr10_+_5689510 | 3.06 |
ENSDART00000183217
ENSDART00000172632 |
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr8_+_23165749 | 3.04 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr23_-_28141419 | 2.99 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr2_+_6885852 | 2.96 |
ENSDART00000016607
|
rgs5b
|
regulator of G protein signaling 5b |
| chr18_-_38088099 | 2.92 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr8_+_28066063 | 2.88 |
ENSDART00000078533
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr17_+_10566490 | 2.86 |
ENSDART00000144408
ENSDART00000137469 |
mgaa
|
MGA, MAX dimerization protein a |
| chr11_+_40649412 | 2.83 |
ENSDART00000043016
ENSDART00000134560 |
slc45a1
|
solute carrier family 45, member 1 |
| chr6_+_40992409 | 2.83 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr17_-_28749640 | 2.82 |
ENSDART00000000948
|
coch
|
coagulation factor C homolog, cochlin (Limulus polyphemus) |
| chr6_+_54538948 | 2.80 |
ENSDART00000149270
|
tulp1b
|
tubby like protein 1b |
| chr8_+_28065803 | 2.79 |
ENSDART00000178481
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr5_-_26181863 | 2.79 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr6_+_49901465 | 2.78 |
ENSDART00000023515
|
chmp4ba
|
charged multivesicular body protein 4Ba |
| chr14_+_34486629 | 2.78 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr6_-_10988741 | 2.78 |
ENSDART00000090709
|
coq7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr1_+_14253118 | 2.75 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr17_+_30448452 | 2.72 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr7_-_48805181 | 2.66 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr18_+_16330025 | 2.64 |
ENSDART00000142353
|
nts
|
neurotensin |
| chr3_-_40301467 | 2.64 |
ENSDART00000055186
|
atp5mf
|
ATP synthase membrane subunit f |
| chr1_+_7546259 | 2.61 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr3_-_42981739 | 2.61 |
ENSDART00000167844
|
mafk
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr13_-_23074238 | 2.59 |
ENSDART00000143210
|
srgn
|
serglycin |
| chr3_+_14463941 | 2.55 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr9_+_11532025 | 2.55 |
ENSDART00000109037
|
cdk5r2b
|
cyclin-dependent kinase 5, regulatory subunit 2b (p39) |
| chr16_-_52540056 | 2.55 |
ENSDART00000188304
|
CR293507.1
|
|
| chr2_+_25929619 | 2.54 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr19_+_10396042 | 2.53 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr5_+_3501859 | 2.52 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr17_+_48174381 | 2.52 |
ENSDART00000191721
ENSDART00000193225 ENSDART00000184542 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr10_+_32683089 | 2.51 |
ENSDART00000063551
|
ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr16_-_16212615 | 2.50 |
ENSDART00000059905
|
upp1
|
uridine phosphorylase 1 |
| chr3_+_16762483 | 2.49 |
ENSDART00000132732
|
tmem86b
|
transmembrane protein 86B |
| chr25_+_33849647 | 2.49 |
ENSDART00000121449
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr25_-_7686201 | 2.49 |
ENSDART00000157267
ENSDART00000155094 |
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr8_+_8845932 | 2.48 |
ENSDART00000112028
|
si:ch211-180f4.1
|
si:ch211-180f4.1 |
| chr21_+_45816030 | 2.48 |
ENSDART00000187056
|
pitx1
|
paired-like homeodomain 1 |
| chr12_+_45200744 | 2.48 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr11_-_17713987 | 2.46 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr6_+_27624023 | 2.45 |
ENSDART00000147789
|
slco2a1
|
solute carrier organic anion transporter family, member 2A1 |
| chr2_-_37478418 | 2.44 |
ENSDART00000146103
|
dapk3
|
death-associated protein kinase 3 |
| chr7_+_23907692 | 2.41 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr9_-_18742704 | 2.40 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr8_-_53649294 | 2.39 |
ENSDART00000158259
|
lrtm1
|
leucine rich repeats and transmembrane domains 1 |
| chr20_+_30490682 | 2.37 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr2_-_27774783 | 2.36 |
ENSDART00000161864
|
XKR4
|
zgc:123035 |
| chr14_-_7888748 | 2.36 |
ENSDART00000166293
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr17_+_48173908 | 2.35 |
ENSDART00000192065
ENSDART00000125738 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr7_+_48761875 | 2.33 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr17_+_51627209 | 2.32 |
ENSDART00000056886
|
zgc:113142
|
zgc:113142 |
| chr21_+_25054420 | 2.32 |
ENSDART00000065132
|
zgc:171740
|
zgc:171740 |
| chr9_-_23922011 | 2.30 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr1_-_38756870 | 2.30 |
ENSDART00000130324
ENSDART00000148404 |
gpm6ab
|
glycoprotein M6Ab |
| chr2_+_9821757 | 2.29 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr14_-_2318590 | 2.27 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr25_-_27564205 | 2.27 |
ENSDART00000157319
|
hyal4
|
hyaluronoglucosaminidase 4 |
| chr24_-_17444067 | 2.26 |
ENSDART00000155843
|
cntnap2a
|
contactin associated protein like 2a |
| chr14_-_33454595 | 2.23 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr10_-_22249444 | 2.23 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr22_+_997838 | 2.22 |
ENSDART00000149743
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr18_+_10784730 | 2.19 |
ENSDART00000028938
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr22_+_28328156 | 2.18 |
ENSDART00000166714
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr13_+_24552254 | 2.17 |
ENSDART00000147907
|
lgalslb
|
lectin, galactoside-binding-like b |
| chr17_-_26721007 | 2.16 |
ENSDART00000034580
|
calm1a
|
calmodulin 1a |
| chr11_+_25638172 | 2.16 |
ENSDART00000114226
ENSDART00000143677 |
grm6b
|
glutamate receptor, metabotropic 6b |
| chr10_-_15128771 | 2.16 |
ENSDART00000101261
|
spp1
|
secreted phosphoprotein 1 |
| chr6_+_112579 | 2.15 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr21_-_24865454 | 2.13 |
ENSDART00000142907
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr3_+_15271943 | 2.13 |
ENSDART00000141752
|
asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr25_-_8030113 | 2.12 |
ENSDART00000104674
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr2_+_20332044 | 2.12 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr20_-_34801181 | 2.12 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr5_+_9360394 | 2.10 |
ENSDART00000124642
|
FP236810.2
|
|
| chr8_+_44714336 | 2.09 |
ENSDART00000145801
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr25_+_13620555 | 2.09 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr12_-_19103490 | 2.08 |
ENSDART00000060561
|
csdc2a
|
cold shock domain containing C2, RNA binding a |
| chr6_-_31348999 | 2.07 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr4_+_13733838 | 2.06 |
ENSDART00000067166
ENSDART00000133157 |
cntn1b
|
contactin 1b |
| chr19_-_12648122 | 2.06 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr7_+_15659280 | 2.03 |
ENSDART00000173414
|
mef2ab
|
myocyte enhancer factor 2ab |
| chr6_-_40063359 | 2.03 |
ENSDART00000157119
ENSDART00000059636 ENSDART00000156385 |
chchd4b
|
coiled-coil-helix-coiled-coil-helix domain containing 4b |
| chr22_-_600016 | 2.03 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr22_+_18786797 | 2.01 |
ENSDART00000141864
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr10_-_39011514 | 2.00 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr14_-_31080183 | 1.99 |
ENSDART00000173282
|
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr2_-_11912347 | 1.98 |
ENSDART00000023851
|
abhd3
|
abhydrolase domain containing 3 |
| chr7_-_72605673 | 1.97 |
ENSDART00000123887
|
MAPK8IP1 (1 of many)
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr11_-_7320211 | 1.96 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr1_-_54765262 | 1.95 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr10_+_252425 | 1.93 |
ENSDART00000059478
|
lrrc32
|
leucine rich repeat containing 32 |
| chr6_+_27146671 | 1.93 |
ENSDART00000156792
|
kif1aa
|
kinesin family member 1Aa |
| chr20_+_3108597 | 1.93 |
ENSDART00000133435
|
CEP170B (1 of many)
|
si:ch73-212j7.1 |
| chr6_+_36795225 | 1.93 |
ENSDART00000171504
|
si:ch73-29l19.1
|
si:ch73-29l19.1 |
| chr24_-_28711176 | 1.92 |
ENSDART00000105753
|
olfm3a
|
olfactomedin 3a |
| chr16_+_4055331 | 1.92 |
ENSDART00000128978
|
CR391998.1
|
|
| chr1_-_46632948 | 1.91 |
ENSDART00000148893
ENSDART00000053232 |
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr7_-_32598383 | 1.90 |
ENSDART00000111055
|
kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr1_+_49266886 | 1.89 |
ENSDART00000137179
|
caly
|
calcyon neuron-specific vesicular protein |
| chr25_-_37084032 | 1.88 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr18_+_17663898 | 1.87 |
ENSDART00000021213
|
cpne2
|
copine II |
| chr6_-_51101834 | 1.86 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr17_+_8184649 | 1.86 |
ENSDART00000091818
|
tulp4b
|
tubby like protein 4b |
| chr14_-_46238186 | 1.86 |
ENSDART00000173245
|
si:ch211-113d11.6
|
si:ch211-113d11.6 |
| chr25_-_23052707 | 1.85 |
ENSDART00000024633
|
dusp8a
|
dual specificity phosphatase 8a |
| chr16_+_14029283 | 1.85 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr23_-_637347 | 1.83 |
ENSDART00000132175
|
l1camb
|
L1 cell adhesion molecule, paralog b |
| chr2_-_42415902 | 1.83 |
ENSDART00000142489
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr19_+_3849378 | 1.81 |
ENSDART00000166218
ENSDART00000159228 |
oscp1a
|
organic solute carrier partner 1a |
| chr5_-_23999777 | 1.81 |
ENSDART00000085969
|
map7d2a
|
MAP7 domain containing 2a |
| chr2_-_27775236 | 1.80 |
ENSDART00000187983
|
XKR4
|
zgc:123035 |
| chr14_+_5936996 | 1.80 |
ENSDART00000097144
ENSDART00000126777 |
kctd8
|
potassium channel tetramerization domain containing 8 |
| chr7_+_13382852 | 1.78 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr12_+_4036409 | 1.76 |
ENSDART00000106650
|
zgc:123217
|
zgc:123217 |
| chr25_-_19134489 | 1.75 |
ENSDART00000193629
|
abhd2b
|
abhydrolase domain containing 2b |
| chr7_-_28413224 | 1.74 |
ENSDART00000076502
|
rerglb
|
RERG/RAS-like b |
| chr21_+_22630297 | 1.73 |
ENSDART00000147175
|
si:dkeyp-69c1.7
|
si:dkeyp-69c1.7 |
| chr2_+_21090317 | 1.73 |
ENSDART00000109568
ENSDART00000139633 |
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr4_-_14915268 | 1.73 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr1_-_40227166 | 1.72 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr11_-_44876005 | 1.72 |
ENSDART00000192006
|
opn6a
|
opsin 6, group member a |
| chr14_-_11430566 | 1.70 |
ENSDART00000137154
ENSDART00000091158 |
irg1l
|
immunoresponsive gene 1, like |
| chr4_+_26496489 | 1.70 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr3_-_49504023 | 1.70 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr7_-_20241346 | 1.70 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr7_+_48761646 | 1.67 |
ENSDART00000017467
|
acana
|
aggrecan a |
| chr17_-_43012390 | 1.67 |
ENSDART00000155615
|
atg2b
|
autophagy related 2B |
| chr10_+_29698467 | 1.67 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr4_+_73973242 | 1.67 |
ENSDART00000182529
|
LO018181.1
|
|
| chr13_+_12175724 | 1.67 |
ENSDART00000166053
|
gabrg1
|
gamma-aminobutyric acid type A receptor gamma1 subunit |
| chr24_-_30091937 | 1.67 |
ENSDART00000148249
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr8_-_5220125 | 1.66 |
ENSDART00000035676
|
bnip3la
|
BCL2 interacting protein 3 like a |
| chr21_+_22630627 | 1.65 |
ENSDART00000193092
|
si:dkeyp-69c1.7
|
si:dkeyp-69c1.7 |
| chr1_-_51606552 | 1.64 |
ENSDART00000130828
|
cnrip1a
|
cannabinoid receptor interacting protein 1a |
| chr7_-_35515931 | 1.64 |
ENSDART00000193324
|
irx6a
|
iroquois homeobox 6a |
| chr7_-_35516251 | 1.64 |
ENSDART00000045628
|
irx6a
|
iroquois homeobox 6a |
| chr21_-_275377 | 1.64 |
ENSDART00000157509
|
rln1
|
relaxin 1 |
| chr1_-_46924801 | 1.63 |
ENSDART00000142560
|
pdxkb
|
pyridoxal (pyridoxine, vitamin B6) kinase b |
| chr2_-_37477654 | 1.63 |
ENSDART00000193921
|
dapk3
|
death-associated protein kinase 3 |
| chr1_-_22861348 | 1.61 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr18_-_46063773 | 1.61 |
ENSDART00000078561
|
si:ch73-262h23.4
|
si:ch73-262h23.4 |
| chr19_-_12648408 | 1.61 |
ENSDART00000103692
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr20_+_34913069 | 1.61 |
ENSDART00000007584
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr9_-_4598883 | 1.61 |
ENSDART00000171927
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr7_-_39203799 | 1.60 |
ENSDART00000173727
|
chrm4a
|
cholinergic receptor, muscarinic 4a |
| chr23_+_28582865 | 1.59 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr23_+_21544227 | 1.59 |
ENSDART00000140253
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr17_+_33313566 | 1.59 |
ENSDART00000045040
|
si:ch211-132f19.7
|
si:ch211-132f19.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of junba+junbb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0097623 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 1.3 | 6.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 1.2 | 4.8 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.9 | 2.8 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.9 | 2.7 | GO:0097435 | fibril organization(GO:0097435) |
| 0.8 | 3.1 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.7 | 3.7 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.7 | 2.8 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.7 | 9.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.7 | 4.0 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.7 | 2.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.6 | 2.6 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.6 | 1.9 | GO:0032263 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.6 | 3.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 1.6 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.5 | 2.6 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.5 | 3.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.5 | 2.0 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.5 | 7.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.5 | 1.4 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.5 | 3.3 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.5 | 4.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.5 | 2.3 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.4 | 1.8 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.4 | 5.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.4 | 2.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.4 | 3.7 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.4 | 1.6 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.4 | 2.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.4 | 2.6 | GO:1903306 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.4 | 1.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.3 | 3.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.3 | 1.4 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.3 | 2.3 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 3.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.3 | 5.2 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.3 | 6.9 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.3 | 1.5 | GO:0072104 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.3 | 1.4 | GO:0031670 | cellular response to nutrient(GO:0031670) negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) cellular response to vitamin(GO:0071295) cellular response to vitamin D(GO:0071305) |
| 0.3 | 2.5 | GO:0071326 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.3 | 0.8 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.3 | 4.0 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 3.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.3 | 1.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 3.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 2.5 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.2 | 0.7 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.2 | 2.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.2 | 1.7 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.2 | 3.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.2 | 0.7 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.2 | 2.1 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) single-organism membrane invagination(GO:1902534) |
| 0.2 | 0.7 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 0.9 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.2 | 3.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 3.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.2 | 0.9 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.2 | 2.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.2 | 3.1 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.2 | 0.6 | GO:0019677 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.2 | 1.6 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.2 | 4.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.2 | 1.7 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.2 | 3.3 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.2 | 1.4 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.2 | 2.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 3.2 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.2 | 2.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 1.9 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.2 | 2.0 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.2 | 1.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.2 | 0.6 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 0.6 | GO:0071788 | endoplasmic reticulum tubular network maintenance(GO:0071788) |
| 0.2 | 2.4 | GO:0009583 | phototransduction(GO:0007602) detection of light stimulus(GO:0009583) |
| 0.2 | 2.5 | GO:0071715 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.2 | 5.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 3.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 1.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 4.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 4.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 1.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.7 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 1.6 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) late endosome to vacuole transport(GO:0045324) |
| 0.1 | 1.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.7 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.8 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 6.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.5 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 3.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.6 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.1 | 0.6 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.2 | GO:0014821 | phasic smooth muscle contraction(GO:0014821) |
| 0.1 | 1.1 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.1 | 2.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.8 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 3.8 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 2.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 1.0 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 1.0 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.5 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 1.1 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 2.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 4.5 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 0.9 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.7 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.7 | GO:0050856 | regulation of T cell receptor signaling pathway(GO:0050856) |
| 0.1 | 3.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.5 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 2.6 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 0.5 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.8 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.1 | 0.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.6 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.9 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 4.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 1.9 | GO:0030168 | platelet activation(GO:0030168) |
| 0.1 | 1.5 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.1 | 0.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 1.8 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 1.0 | GO:0072028 | nephron morphogenesis(GO:0072028) |
| 0.1 | 5.4 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 2.2 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 1.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.4 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.1 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 1.1 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 3.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 1.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 2.5 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.1 | 0.5 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 7.7 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 5.4 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 | 4.0 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.1 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.7 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 1.1 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 1.6 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 1.5 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 5.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 1.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.5 | GO:1903307 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) positive regulation of regulated secretory pathway(GO:1903307) |
| 0.1 | 1.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:0032988 | spliceosomal complex disassembly(GO:0000390) ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.3 | GO:0006574 | thymine catabolic process(GO:0006210) valine catabolic process(GO:0006574) thymine metabolic process(GO:0019859) |
| 0.0 | 9.5 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 3.3 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.6 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.5 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.5 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 5.7 | GO:0043068 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.9 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 1.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.3 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.3 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.7 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 1.4 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 1.6 | GO:0006171 | cAMP biosynthetic process(GO:0006171) cAMP metabolic process(GO:0046058) |
| 0.0 | 0.2 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 1.8 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 4.1 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.6 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) regulation of polysaccharide biosynthetic process(GO:0032885) |
| 0.0 | 1.7 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 2.1 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 1.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.6 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 6.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.8 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.7 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.7 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.4 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.6 | GO:0045727 | positive regulation of cellular amide metabolic process(GO:0034250) positive regulation of translation(GO:0045727) |
| 0.0 | 0.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.8 | GO:0000302 | response to reactive oxygen species(GO:0000302) |
| 0.0 | 0.8 | GO:0060401 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 3.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.9 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 1.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.8 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.4 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.2 | GO:0015800 | C4-dicarboxylate transport(GO:0015740) acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 1.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 1.0 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.5 | GO:0001503 | ossification(GO:0001503) |
| 0.0 | 1.3 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.3 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.4 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.4 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.6 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.4 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.5 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 11.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.7 | 5.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.5 | 2.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.5 | 5.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 1.7 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.4 | 3.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 2.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 4.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.3 | 2.8 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 3.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 2.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.3 | 4.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 5.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 0.7 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 1.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 2.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 0.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 0.7 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 6.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 11.9 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 2.8 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 1.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 4.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 6.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 4.1 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.5 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.7 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 2.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.8 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 3.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 2.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 3.7 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 2.7 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 1.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.9 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.7 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 1.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.0 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 4.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 6.4 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 4.2 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.0 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 2.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 1.0 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 2.8 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 45.2 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 6.2 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 23.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 1.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.1 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 3.0 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.8 | 3.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.8 | 3.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.7 | 6.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.7 | 2.8 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.7 | 3.4 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.7 | 2.0 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.6 | 3.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.6 | 1.9 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.6 | 3.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 3.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.5 | 2.7 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.5 | 3.7 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.5 | 2.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.5 | 7.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.5 | 6.9 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.4 | 4.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 2.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 3.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.4 | 2.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.4 | 1.9 | GO:0030955 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.4 | 4.9 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.3 | 1.7 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.3 | 1.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 2.8 | GO:0015157 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.3 | 1.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 1.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 8.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 1.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 1.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 2.5 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.3 | 4.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.3 | 1.4 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.3 | 3.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 0.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.3 | 3.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 0.8 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.3 | 10.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 9.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 2.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 7.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 5.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.2 | 2.9 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 3.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.2 | 1.5 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.2 | 0.7 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 5.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 1.4 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.2 | 1.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 0.7 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.2 | 2.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 1.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 0.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 1.8 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.2 | 2.5 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.2 | 0.6 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.2 | 3.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 5.4 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.1 | 1.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 2.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 2.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.0 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.6 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.9 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.5 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.1 | 9.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 6.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 1.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.9 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 5.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.7 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 1.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.9 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 3.2 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 1.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 6.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 4.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 8.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 3.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 2.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 2.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.2 | GO:0016917 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.1 | 0.4 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 2.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 1.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 2.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 1.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.8 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.8 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.1 | 1.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 2.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 1.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 8.2 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 1.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 6.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 4.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 1.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 1.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 2.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.2 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.4 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 6.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 1.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.2 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.5 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.6 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 2.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.6 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 5.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 11.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 5.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.1 | GO:0004407 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.0 | 1.6 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 2.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 3.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.3 | 4.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 3.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 4.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 4.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 8.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 0.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 5.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 2.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 0.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 1.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 2.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 2.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 2.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.3 | 9.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 3.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 2.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 2.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 2.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 1.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 5.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 6.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.6 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 2.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 0.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.5 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 1.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 2.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.5 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.8 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |