Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
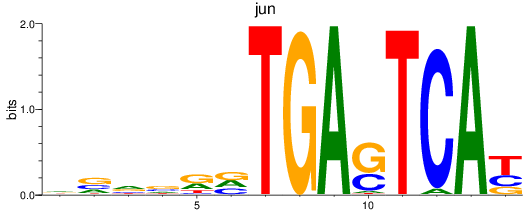
Results for jun
Z-value: 1.00
Transcription factors associated with jun
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
jun
|
ENSDARG00000043531 | Jun proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| jun | dr11_v1_chr20_+_15552657_15552657 | 0.92 | 3.8e-08 | Click! |
Activity profile of jun motif
Sorted Z-values of jun motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_48359259 | 3.33 |
ENSDART00000167353
|
sgo1
|
shugoshin 1 |
| chr5_-_20195350 | 2.49 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr20_-_26531850 | 2.40 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr20_-_26532167 | 2.39 |
ENSDART00000061914
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr20_+_28364742 | 2.37 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr16_+_27543893 | 2.27 |
ENSDART00000182421
|
si:ch211-197h24.6
|
si:ch211-197h24.6 |
| chr20_-_25626198 | 2.24 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr23_-_21471022 | 1.90 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr11_-_24681292 | 1.82 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr23_+_35714574 | 1.50 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr12_+_25945560 | 1.49 |
ENSDART00000109799
|
mmrn2b
|
multimerin 2b |
| chr10_+_22771176 | 1.45 |
ENSDART00000192046
|
tmem88a
|
transmembrane protein 88 a |
| chr24_-_31090948 | 1.40 |
ENSDART00000176799
|
hccsb
|
holocytochrome c synthase b |
| chr10_+_38593645 | 1.38 |
ENSDART00000011573
|
mmp13a
|
matrix metallopeptidase 13a |
| chr14_-_17588345 | 1.35 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr5_-_30615901 | 1.34 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr22_+_16497670 | 1.32 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr16_-_21785261 | 1.30 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr11_+_24251141 | 1.30 |
ENSDART00000182684
|
pnp4a
|
purine nucleoside phosphorylase 4a |
| chr9_-_48370645 | 1.25 |
ENSDART00000140185
|
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr22_+_24220937 | 1.23 |
ENSDART00000165380
|
uchl5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr8_-_2616326 | 1.19 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr16_+_23403602 | 1.17 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr3_-_26017592 | 1.16 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr3_-_26017831 | 1.15 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr6_-_21534301 | 1.11 |
ENSDART00000126186
|
psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr24_+_31361407 | 1.10 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr19_+_37120491 | 1.08 |
ENSDART00000032341
|
pef1
|
penta-EF-hand domain containing 1 |
| chr25_-_13408760 | 1.07 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr9_+_41024973 | 1.05 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr1_-_55116453 | 1.04 |
ENSDART00000142348
|
sertad2a
|
SERTA domain containing 2a |
| chr10_+_32050906 | 1.04 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr19_-_42573219 | 1.04 |
ENSDART00000126021
ENSDART00000133695 ENSDART00000131558 |
zgc:103438
|
zgc:103438 |
| chr19_+_15441022 | 1.04 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr4_-_13902188 | 1.04 |
ENSDART00000032805
|
gxylt1b
|
glucoside xylosyltransferase 1b |
| chr6_+_44197348 | 1.02 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr6_+_44197099 | 1.02 |
ENSDART00000124168
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr2_-_37837472 | 1.02 |
ENSDART00000165347
|
mettl17
|
methyltransferase like 17 |
| chr23_+_24124684 | 1.01 |
ENSDART00000144478
|
si:dkey-21o19.2
|
si:dkey-21o19.2 |
| chr13_-_9213207 | 1.00 |
ENSDART00000139861
ENSDART00000140524 |
HTRA2 (1 of many)
|
si:dkey-33c12.11 |
| chr5_+_44804791 | 1.00 |
ENSDART00000122288
|
ctsla
|
cathepsin La |
| chr4_+_16715267 | 1.00 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr7_+_50395856 | 0.99 |
ENSDART00000032324
|
hddc3
|
HD domain containing 3 |
| chr25_-_12824939 | 0.99 |
ENSDART00000169126
|
uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr19_+_15443063 | 0.97 |
ENSDART00000151732
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr2_+_30379650 | 0.96 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr14_+_46216703 | 0.96 |
ENSDART00000136045
ENSDART00000142317 |
mgst2
|
microsomal glutathione S-transferase 2 |
| chr8_+_50534948 | 0.96 |
ENSDART00000174435
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr2_+_35612621 | 0.95 |
ENSDART00000143082
|
si:dkey-4i23.5
|
si:dkey-4i23.5 |
| chr19_-_7420867 | 0.94 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr5_+_24089334 | 0.94 |
ENSDART00000183748
|
tp53
|
tumor protein p53 |
| chr4_-_6809323 | 0.94 |
ENSDART00000099467
|
ifrd1
|
interferon-related developmental regulator 1 |
| chr11_+_11267829 | 0.93 |
ENSDART00000026814
ENSDART00000173346 ENSDART00000151926 |
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr19_+_24488403 | 0.93 |
ENSDART00000052421
|
txnipa
|
thioredoxin interacting protein a |
| chr5_+_34549845 | 0.92 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr13_+_24280380 | 0.91 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr5_+_44805269 | 0.91 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr7_-_28696556 | 0.91 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr8_+_28695914 | 0.89 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr3_-_39648021 | 0.88 |
ENSDART00000055171
|
grapa
|
GRB2-related adaptor protein a |
| chr10_+_38610741 | 0.87 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr1_+_46598764 | 0.87 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr8_-_22288004 | 0.87 |
ENSDART00000100042
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr3_-_30488063 | 0.85 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr3_-_21062706 | 0.85 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr4_+_77948970 | 0.84 |
ENSDART00000149636
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr3_+_27027781 | 0.84 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr2_-_39675829 | 0.83 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr14_+_30774032 | 0.83 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr14_+_30774515 | 0.82 |
ENSDART00000191666
|
atl3
|
atlastin 3 |
| chr3_+_32118670 | 0.81 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr10_+_31222656 | 0.81 |
ENSDART00000140988
ENSDART00000143387 |
tmem218
|
transmembrane protein 218 |
| chr7_-_22941472 | 0.81 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr20_+_25625872 | 0.81 |
ENSDART00000078385
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr1_+_59088205 | 0.81 |
ENSDART00000150649
ENSDART00000100197 |
zgc:173915
|
zgc:173915 |
| chr21_+_20901505 | 0.81 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr23_+_36130883 | 0.80 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr23_-_36446307 | 0.80 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr25_+_30196039 | 0.79 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr19_-_27564458 | 0.79 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr21_+_8341774 | 0.78 |
ENSDART00000129749
ENSDART00000055325 ENSDART00000133804 |
psmb7
|
proteasome subunit beta 7 |
| chr18_+_7543347 | 0.78 |
ENSDART00000103467
|
arf5
|
ADP-ribosylation factor 5 |
| chr25_-_12824656 | 0.78 |
ENSDART00000171801
|
uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr25_+_32474031 | 0.78 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr6_-_35439406 | 0.77 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr4_+_5317483 | 0.76 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr25_+_2282265 | 0.76 |
ENSDART00000076417
|
cdkn1bb
|
cyclin-dependent kinase inhibitor 1Bb |
| chr9_-_2594410 | 0.74 |
ENSDART00000188306
ENSDART00000164276 |
sp9
|
sp9 transcription factor |
| chr10_-_210703 | 0.74 |
ENSDART00000059476
|
psmg1
|
proteasome (prosome, macropain) assembly chaperone 1 |
| chr2_+_48288461 | 0.73 |
ENSDART00000141495
|
hes6
|
hes family bHLH transcription factor 6 |
| chr8_-_30204650 | 0.72 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr21_-_21178410 | 0.72 |
ENSDART00000185277
ENSDART00000141341 ENSDART00000145872 ENSDART00000079678 |
ftsj1
|
FtsJ RNA methyltransferase homolog 1 |
| chr5_+_44805028 | 0.72 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr19_+_48019070 | 0.71 |
ENSDART00000158269
|
UBE2M
|
si:ch1073-205c8.3 |
| chr6_-_43449013 | 0.71 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr7_+_69019851 | 0.70 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr10_+_16225117 | 0.69 |
ENSDART00000169885
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr2_+_17524278 | 0.69 |
ENSDART00000165633
|
pimr196
|
Pim proto-oncogene, serine/threonine kinase, related 196 |
| chr19_+_48018802 | 0.68 |
ENSDART00000161339
ENSDART00000166978 |
UBE2M
|
si:ch1073-205c8.3 |
| chr25_+_14697247 | 0.68 |
ENSDART00000180747
|
mpped2
|
metallophosphoesterase domain containing 2b |
| chr12_-_27212596 | 0.68 |
ENSDART00000153101
|
psme3
|
proteasome activator subunit 3 |
| chr1_-_58913813 | 0.67 |
ENSDART00000056494
|
zgc:171687
|
zgc:171687 |
| chr1_+_57041549 | 0.67 |
ENSDART00000152198
|
si:ch211-1f22.16
|
si:ch211-1f22.16 |
| chr25_+_29474583 | 0.67 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr8_-_25338709 | 0.67 |
ENSDART00000131616
|
atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr8_+_4434422 | 0.66 |
ENSDART00000145184
|
srrd
|
SRR1 domain containing |
| chr7_-_35432901 | 0.66 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr9_+_33216945 | 0.66 |
ENSDART00000134029
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr19_-_31035155 | 0.66 |
ENSDART00000161882
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr7_-_44704910 | 0.66 |
ENSDART00000037850
|
dync1li2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr8_-_20243389 | 0.66 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr1_+_59090972 | 0.65 |
ENSDART00000171497
|
mfap4
|
microfibril associated protein 4 |
| chr5_+_37087583 | 0.65 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr4_-_16412084 | 0.65 |
ENSDART00000188460
|
dcn
|
decorin |
| chr11_+_13630107 | 0.65 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr10_+_37145007 | 0.65 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr1_+_14253118 | 0.65 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr19_-_27570333 | 0.64 |
ENSDART00000146562
ENSDART00000179060 |
si:dkeyp-46h3.5
si:dkeyp-46h3.8
|
si:dkeyp-46h3.5 si:dkeyp-46h3.8 |
| chr2_+_15100742 | 0.63 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr7_+_22688781 | 0.63 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr8_+_39634114 | 0.63 |
ENSDART00000144293
|
msi1
|
musashi RNA-binding protein 1 |
| chr14_-_26436951 | 0.63 |
ENSDART00000140173
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr7_-_26262978 | 0.62 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr1_+_41849152 | 0.62 |
ENSDART00000053685
|
smox
|
spermine oxidase |
| chr16_+_46410520 | 0.62 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr3_-_30248277 | 0.61 |
ENSDART00000133516
ENSDART00000077029 |
emc10
|
ER membrane protein complex subunit 10 |
| chr3_-_34034498 | 0.61 |
ENSDART00000151145
|
ighv11-2
|
immunoglobulin heavy variable 11-2 |
| chr1_+_59090743 | 0.61 |
ENSDART00000100199
|
mfap4
|
microfibril associated protein 4 |
| chr15_-_28223757 | 0.61 |
ENSDART00000110969
ENSDART00000138401 |
scarf1
|
scavenger receptor class F, member 1 |
| chr18_-_13360106 | 0.61 |
ENSDART00000091512
|
cmip
|
c-Maf inducing protein |
| chr7_+_41147732 | 0.60 |
ENSDART00000185388
|
puf60b
|
poly-U binding splicing factor b |
| chr2_+_25378457 | 0.60 |
ENSDART00000089108
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr14_+_33882973 | 0.60 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr19_-_38539670 | 0.60 |
ENSDART00000136775
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr11_+_29790626 | 0.60 |
ENSDART00000067822
|
dynlt3
|
dynein, light chain, Tctex-type 3 |
| chr6_-_33924883 | 0.60 |
ENSDART00000132762
ENSDART00000148142 ENSDART00000142213 |
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr14_+_15768942 | 0.60 |
ENSDART00000158998
|
ergic1
|
endoplasmic reticulum-golgi intermediate compartment 1 |
| chr14_-_6936731 | 0.59 |
ENSDART00000162196
|
clk4a
|
CDC-like kinase 4a |
| chr24_-_24797455 | 0.59 |
ENSDART00000138741
|
pde7a
|
phosphodiesterase 7A |
| chr20_-_42972599 | 0.59 |
ENSDART00000100751
|
pomcb
|
proopiomelanocortin b |
| chr1_+_59073436 | 0.59 |
ENSDART00000161642
|
MFAP4 (1 of many)
|
si:zfos-2330d3.3 |
| chr5_-_25582721 | 0.58 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr7_+_38680034 | 0.58 |
ENSDART00000171691
ENSDART00000007913 |
psmc3
|
proteasome 26S subunit, ATPase 3 |
| chr21_-_21020708 | 0.57 |
ENSDART00000064032
|
eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr2_-_38080075 | 0.57 |
ENSDART00000056544
|
tox4a
|
TOX high mobility group box family member 4 a |
| chr20_+_35445462 | 0.57 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr23_+_28377360 | 0.57 |
ENSDART00000014983
ENSDART00000128831 ENSDART00000135178 ENSDART00000138621 |
zgc:153867
|
zgc:153867 |
| chr16_-_45225520 | 0.57 |
ENSDART00000158855
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr25_+_10485103 | 0.56 |
ENSDART00000104576
|
psmd13
|
proteasome 26S subunit, non-ATPase 13 |
| chr13_-_17729474 | 0.56 |
ENSDART00000013011
|
vdac2
|
voltage-dependent anion channel 2 |
| chr3_+_46762703 | 0.56 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr23_+_28128453 | 0.55 |
ENSDART00000182618
|
c1galt1a
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1a |
| chr25_-_11378623 | 0.55 |
ENSDART00000166586
|
enc2
|
ectodermal-neural cortex 2 |
| chr1_+_12766351 | 0.55 |
ENSDART00000165785
|
pcdh10a
|
protocadherin 10a |
| chr23_+_42336084 | 0.55 |
ENSDART00000158959
ENSDART00000161812 |
cyp2aa7
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr2_-_39759059 | 0.55 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr16_+_42829735 | 0.55 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr11_-_22371105 | 0.55 |
ENSDART00000146873
|
tmem183a
|
transmembrane protein 183A |
| chr19_-_43757568 | 0.54 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr11_-_13126969 | 0.54 |
ENSDART00000169052
ENSDART00000190120 |
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr20_-_43723860 | 0.54 |
ENSDART00000122051
|
mixl1
|
Mix paired-like homeobox |
| chr8_+_50531709 | 0.53 |
ENSDART00000193352
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr10_+_187760 | 0.53 |
ENSDART00000161007
ENSDART00000160651 |
ets2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr1_-_22726233 | 0.53 |
ENSDART00000140920
|
prom1b
|
prominin 1 b |
| chr7_-_44963154 | 0.53 |
ENSDART00000073735
|
rrad
|
Ras-related associated with diabetes |
| chr5_+_42064144 | 0.53 |
ENSDART00000035235
|
si:ch211-202a12.4
|
si:ch211-202a12.4 |
| chr8_-_18667693 | 0.52 |
ENSDART00000100516
|
stap2b
|
signal transducing adaptor family member 2b |
| chr4_-_28958601 | 0.52 |
ENSDART00000111294
|
zgc:174315
|
zgc:174315 |
| chr17_+_12942021 | 0.52 |
ENSDART00000192514
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr1_-_5746030 | 0.52 |
ENSDART00000150863
|
nrp2a
|
neuropilin 2a |
| chr5_-_55623443 | 0.52 |
ENSDART00000005671
ENSDART00000176341 |
hnrpkl
|
heterogeneous nuclear ribonucleoprotein K, like |
| chr16_+_37876779 | 0.51 |
ENSDART00000140148
|
si:ch211-198c19.1
|
si:ch211-198c19.1 |
| chr11_+_11267493 | 0.51 |
ENSDART00000148425
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr13_-_17723417 | 0.51 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr4_-_4535189 | 0.50 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr2_-_37862380 | 0.50 |
ENSDART00000186005
|
si:ch211-284o19.8
|
si:ch211-284o19.8 |
| chr6_-_18976168 | 0.50 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr11_+_2416064 | 0.50 |
ENSDART00000067117
|
ube2v1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr19_-_32641725 | 0.50 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr19_-_31035325 | 0.49 |
ENSDART00000147504
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr8_+_32406885 | 0.49 |
ENSDART00000167600
|
epgn
|
epithelial mitogen homolog (mouse) |
| chr25_+_7423770 | 0.49 |
ENSDART00000155458
|
ubap1la
|
ubiquitin associated protein 1-like a |
| chr1_+_44340124 | 0.49 |
ENSDART00000160696
|
CT033803.2
|
|
| chr11_+_14057605 | 0.49 |
ENSDART00000166262
|
grin3bb
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3Bb |
| chr20_-_21672970 | 0.49 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr25_+_18583877 | 0.49 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr13_+_15800742 | 0.49 |
ENSDART00000146234
|
apopt1
|
apoptogenic 1, mitochondrial |
| chr9_+_33217243 | 0.48 |
ENSDART00000053061
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr1_+_58677434 | 0.48 |
ENSDART00000158488
|
LO017784.1
|
|
| chr7_-_52963493 | 0.48 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr1_-_22861348 | 0.48 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr23_+_29570386 | 0.48 |
ENSDART00000193192
|
lzic
|
leucine zipper and CTNNBIP1 domain containing |
| chr25_-_11026907 | 0.48 |
ENSDART00000156846
|
mespbb
|
mesoderm posterior bb |
| chr12_+_20587179 | 0.48 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr9_+_29323163 | 0.48 |
ENSDART00000184908
|
oxgr1b
|
oxoglutarate (alpha-ketoglutarate) receptor 1b |
| chr23_-_5783421 | 0.48 |
ENSDART00000131521
ENSDART00000019455 |
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr1_-_30762264 | 0.47 |
ENSDART00000085454
|
dis3
|
DIS3 exosome endoribonuclease and 3'-5' exoribonuclease |
| chr17_+_33496067 | 0.47 |
ENSDART00000184724
|
pth2
|
parathyroid hormone 2 |
| chr8_-_22288258 | 0.47 |
ENSDART00000140978
ENSDART00000100046 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr1_+_44249616 | 0.47 |
ENSDART00000164173
|
si:ch211-165a10.10
|
si:ch211-165a10.10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of jun
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.8 | 2.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.8 | 2.3 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.6 | 2.5 | GO:0055130 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.5 | 1.5 | GO:0061317 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.3 | 4.8 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.3 | 0.9 | GO:0043525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.3 | 3.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.3 | 0.8 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.3 | 1.4 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.3 | 1.1 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.3 | 0.8 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.2 | 1.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 0.6 | GO:0002631 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.2 | 1.0 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 0.6 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 2.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 1.1 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.2 | 0.9 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.7 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 1.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.7 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 1.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.8 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.5 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.7 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.8 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.6 | GO:0045628 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 2.5 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.1 | 0.4 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 0.6 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 1.9 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.4 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.1 | 0.8 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 0.5 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.8 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.5 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 0.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.2 | GO:0032816 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.3 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.1 | 0.2 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 1.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.5 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.2 | GO:0043084 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.1 | 0.7 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.5 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 1.8 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.1 | 2.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 0.4 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 0.6 | GO:0030719 | P granule organization(GO:0030719) |
| 0.1 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.6 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 1.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.9 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.9 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.5 | GO:0021707 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.3 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.0 | 0.7 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.5 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.4 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.6 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.2 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.0 | 0.8 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.3 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.5 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.4 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.3 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.4 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.7 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.5 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.4 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.5 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.0 | 0.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.7 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.5 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.4 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.4 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.6 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.5 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.2 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.7 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.5 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.4 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0036135 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.0 | 0.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.3 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 1.0 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:1903707 | negative regulation of hemopoiesis(GO:1903707) |
| 0.0 | 0.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 1.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 3.5 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.5 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.3 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.6 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.6 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.4 | GO:0030278 | regulation of ossification(GO:0030278) |
| 0.0 | 0.3 | GO:0032355 | response to estradiol(GO:0032355) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.4 | 1.8 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.3 | 1.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 1.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 0.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 0.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 1.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.6 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 2.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 1.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.0 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.5 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 0.5 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.5 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.2 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.1 | 1.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.5 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 3.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.9 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 2.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 4.4 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.9 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.3 | GO:0005635 | nuclear envelope(GO:0005635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.8 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.6 | 2.5 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.6 | 2.2 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.4 | 1.8 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.4 | 2.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.3 | 1.4 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.3 | 0.8 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.2 | 1.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.2 | 1.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.2 | 1.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 1.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 0.6 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 0.7 | GO:0016504 | peptidase activator activity(GO:0016504) endopeptidase activator activity(GO:0061133) |
| 0.2 | 1.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 0.6 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 1.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 1.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.4 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 2.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.6 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.6 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.1 | 0.8 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 3.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.4 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.7 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.7 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.2 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 0.6 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.4 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 0.2 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.1 | 1.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.4 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.5 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.3 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 1.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.8 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.6 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 4.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.3 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.7 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.4 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 6.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.4 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.8 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.0 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 0.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.8 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 3.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 3.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |