Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
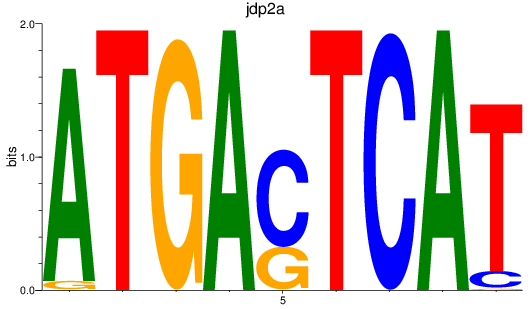
Results for jdp2a
Z-value: 0.85
Transcription factors associated with jdp2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
jdp2a
|
ENSDARG00000040137 | Jun dimerization protein 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| jdp2a | dr11_v1_chr17_-_50220228_50220228 | 0.53 | 1.9e-02 | Click! |
Activity profile of jdp2a motif
Sorted Z-values of jdp2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_21832441 | 5.01 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr8_-_20838342 | 3.49 |
ENSDART00000141345
|
si:ch211-133l5.7
|
si:ch211-133l5.7 |
| chr16_-_36834505 | 3.48 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr13_-_7031033 | 3.42 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr6_-_12588044 | 3.35 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr13_+_23988442 | 3.29 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr8_+_43053519 | 3.20 |
ENSDART00000147178
|
prnpa
|
prion protein a |
| chr16_+_18974064 | 2.98 |
ENSDART00000079248
|
slc6a19b
|
solute carrier family 6 (neutral amino acid transporter), member 19b |
| chr11_-_3552067 | 2.50 |
ENSDART00000163656
|
CAMK2N1
|
si:dkey-33m11.6 |
| chr16_-_13818061 | 2.45 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr6_-_43449013 | 2.38 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr3_+_24986145 | 2.37 |
ENSDART00000055428
|
cbx7a
|
chromobox homolog 7a |
| chr2_+_30379650 | 2.34 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr1_-_43905252 | 2.30 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr22_-_10110959 | 2.26 |
ENSDART00000031005
ENSDART00000147580 |
gls2b
|
glutaminase 2b (liver, mitochondrial) |
| chr6_+_40354424 | 2.25 |
ENSDART00000047416
|
slc4a8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr8_+_24854600 | 2.20 |
ENSDART00000156570
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr7_+_34492744 | 2.19 |
ENSDART00000109635
ENSDART00000173844 |
calml4a
|
calmodulin-like 4a |
| chr3_-_61185746 | 2.17 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr22_-_24297510 | 2.01 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr1_+_45323142 | 2.00 |
ENSDART00000132210
|
emp1
|
epithelial membrane protein 1 |
| chr2_+_47582488 | 1.98 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr1_-_54765262 | 1.94 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr7_-_13381129 | 1.89 |
ENSDART00000164326
|
si:ch73-119p20.1
|
si:ch73-119p20.1 |
| chr24_-_24849091 | 1.87 |
ENSDART00000133649
ENSDART00000038290 |
crhb
|
corticotropin releasing hormone b |
| chr22_+_19247255 | 1.82 |
ENSDART00000144053
|
si:dkey-21e2.10
|
si:dkey-21e2.10 |
| chr1_+_45323400 | 1.80 |
ENSDART00000148906
ENSDART00000132366 |
emp1
|
epithelial membrane protein 1 |
| chr2_+_47581997 | 1.75 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr4_-_69189894 | 1.74 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr2_+_47582681 | 1.72 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr3_+_7617353 | 1.70 |
ENSDART00000165551
|
zgc:109949
|
zgc:109949 |
| chr18_+_21408794 | 1.56 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr9_-_22057658 | 1.53 |
ENSDART00000101944
|
crygmxl1
|
crystallin, gamma MX, like 1 |
| chr8_-_14375890 | 1.53 |
ENSDART00000090306
|
xpr1a
|
xenotropic and polytropic retrovirus receptor 1a |
| chr6_+_49901465 | 1.49 |
ENSDART00000023515
|
chmp4ba
|
charged multivesicular body protein 4Ba |
| chr7_+_20587506 | 1.46 |
ENSDART00000172416
ENSDART00000170633 |
si:dkey-19b23.7
|
si:dkey-19b23.7 |
| chr3_-_58189429 | 1.40 |
ENSDART00000156092
|
si:ch211-256e16.11
|
si:ch211-256e16.11 |
| chr19_-_103289 | 1.39 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr3_+_45401472 | 1.39 |
ENSDART00000156693
|
baiap3
|
BAI1-associated protein 3 |
| chr8_+_10561922 | 1.37 |
ENSDART00000133348
|
fam19a5l
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5-like |
| chr20_-_35512932 | 1.35 |
ENSDART00000137690
|
adgrf3b
|
adhesion G protein-coupled receptor F3b |
| chr7_+_48460239 | 1.31 |
ENSDART00000052113
|
lingo1b
|
leucine rich repeat and Ig domain containing 1b |
| chr1_-_45647846 | 1.30 |
ENSDART00000186881
|
BX511120.1
|
|
| chr8_-_22288258 | 1.29 |
ENSDART00000140978
ENSDART00000100046 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr18_+_23218980 | 1.29 |
ENSDART00000185014
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr19_-_5805923 | 1.25 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr7_-_29534001 | 1.23 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr11_-_24458786 | 1.22 |
ENSDART00000089713
|
mxra8a
|
matrix-remodelling associated 8a |
| chr1_+_31113951 | 1.20 |
ENSDART00000129362
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr7_+_528593 | 1.20 |
ENSDART00000091955
|
nrxn2b
|
neurexin 2b |
| chr10_+_37145007 | 1.18 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr4_+_55273988 | 1.16 |
ENSDART00000163388
|
si:dkey-157e10.8
|
si:dkey-157e10.8 |
| chr21_+_26612777 | 1.15 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr6_+_11250316 | 1.15 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr1_+_6640437 | 1.13 |
ENSDART00000147638
|
si:ch211-93g23.2
|
si:ch211-93g23.2 |
| chr9_+_33154841 | 1.11 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr12_-_10315039 | 1.09 |
ENSDART00000152680
|
pyyb
|
peptide YYb |
| chr23_+_9067131 | 1.08 |
ENSDART00000144533
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr20_-_40319890 | 1.07 |
ENSDART00000075112
|
clvs2
|
clavesin 2 |
| chr20_-_16548912 | 1.02 |
ENSDART00000137601
|
ches1
|
checkpoint suppressor 1 |
| chr19_-_36675023 | 1.02 |
ENSDART00000132471
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr10_-_34772211 | 1.02 |
ENSDART00000145450
ENSDART00000134307 |
dclk1a
|
doublecortin-like kinase 1a |
| chr8_-_13177536 | 1.02 |
ENSDART00000100992
|
zgc:194990
|
zgc:194990 |
| chr7_+_3357973 | 1.01 |
ENSDART00000172853
|
si:ch211-285c6.5
|
si:ch211-285c6.5 |
| chr7_-_60351876 | 1.00 |
ENSDART00000098563
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr7_+_44802353 | 0.99 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| chr23_-_37514493 | 0.98 |
ENSDART00000083267
|
dnajc16
|
DnaJ (Hsp40) homolog, subfamily C, member 16 |
| chr2_+_9822319 | 0.97 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr5_-_54482245 | 0.97 |
ENSDART00000177470
|
fbxw5
|
F-box and WD repeat domain containing 5 |
| chr24_-_21404367 | 0.97 |
ENSDART00000152093
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr13_+_50368668 | 0.96 |
ENSDART00000127610
|
dnajc12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr18_-_9046805 | 0.95 |
ENSDART00000134224
|
grm3
|
glutamate receptor, metabotropic 3 |
| chr21_-_37733571 | 0.95 |
ENSDART00000176214
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr2_-_6519017 | 0.94 |
ENSDART00000181716
|
rgs1
|
regulator of G protein signaling 1 |
| chr19_+_4139065 | 0.94 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr11_-_24681292 | 0.92 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr10_+_4499943 | 0.92 |
ENSDART00000125299
|
plk2a
|
polo-like kinase 2a (Drosophila) |
| chr5_+_52706316 | 0.91 |
ENSDART00000169488
|
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr23_+_39462208 | 0.90 |
ENSDART00000136707
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr25_-_173165 | 0.89 |
ENSDART00000193594
|
CABZ01114053.1
|
|
| chr23_+_39461989 | 0.87 |
ENSDART00000184761
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr5_-_64355227 | 0.87 |
ENSDART00000170787
|
fam78aa
|
family with sequence similarity 78, member Aa |
| chr14_+_34486629 | 0.85 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr2_+_38264964 | 0.85 |
ENSDART00000182068
|
dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr13_-_40141630 | 0.84 |
ENSDART00000181618
ENSDART00000171583 ENSDART00000192780 |
crtac1b
|
cartilage acidic protein 1b |
| chr12_+_7497882 | 0.84 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr5_+_20035284 | 0.81 |
ENSDART00000191808
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr8_-_22288004 | 0.81 |
ENSDART00000100042
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr5_-_57480660 | 0.80 |
ENSDART00000147875
ENSDART00000142776 |
si:ch211-202f5.2
|
si:ch211-202f5.2 |
| chr17_+_45413324 | 0.80 |
ENSDART00000124911
|
ezra
|
ezrin a |
| chr1_+_51312752 | 0.80 |
ENSDART00000063938
|
mast1a
|
microtubule associated serine/threonine kinase 1a |
| chr3_+_16976095 | 0.78 |
ENSDART00000112450
|
cavin1a
|
caveolae associated protein 1a |
| chr12_-_6880694 | 0.77 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr21_+_3093419 | 0.77 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr22_-_16416882 | 0.76 |
ENSDART00000062749
|
cts12
|
cathepsin 12 |
| chr4_-_72643171 | 0.75 |
ENSDART00000130126
|
CABZ01054394.1
|
|
| chr17_+_41302660 | 0.74 |
ENSDART00000059480
|
fosl2
|
fos-like antigen 2 |
| chr20_+_23173710 | 0.74 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr7_-_60351537 | 0.73 |
ENSDART00000159875
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr19_-_31035325 | 0.72 |
ENSDART00000147504
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr24_-_6671549 | 0.71 |
ENSDART00000114700
|
unm_sa821
|
un-named sa821 |
| chr2_+_15100742 | 0.71 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr16_-_12984631 | 0.70 |
ENSDART00000184863
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr25_+_8407892 | 0.69 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr19_-_31035155 | 0.67 |
ENSDART00000161882
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr2_-_49997055 | 0.67 |
ENSDART00000140294
|
si:ch211-106n13.3
|
si:ch211-106n13.3 |
| chr22_+_23359369 | 0.66 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr8_+_5024468 | 0.65 |
ENSDART00000030938
|
adra1aa
|
adrenoceptor alpha 1Aa |
| chr16_+_23403602 | 0.65 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr12_+_47280839 | 0.65 |
ENSDART00000187574
|
RYR2
|
ryanodine receptor 2 |
| chr18_+_14342326 | 0.64 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr13_+_18701906 | 0.64 |
ENSDART00000160779
ENSDART00000136749 |
pdzd7a
|
PDZ domain containing 7a |
| chr8_-_47818282 | 0.63 |
ENSDART00000137149
|
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr1_+_12766351 | 0.63 |
ENSDART00000165785
|
pcdh10a
|
protocadherin 10a |
| chr6_-_33685325 | 0.62 |
ENSDART00000181883
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr8_-_11170114 | 0.61 |
ENSDART00000133532
|
si:ch211-204d2.4
|
si:ch211-204d2.4 |
| chr17_-_7861219 | 0.61 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr3_+_39922602 | 0.60 |
ENSDART00000193937
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr19_+_33701366 | 0.59 |
ENSDART00000162517
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr7_+_5529967 | 0.58 |
ENSDART00000142709
|
si:dkeyp-67a8.4
|
si:dkeyp-67a8.4 |
| chr8_+_48965767 | 0.57 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr2_+_9821757 | 0.56 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr2_+_21229909 | 0.56 |
ENSDART00000046127
|
vipr1a
|
vasoactive intestinal peptide receptor 1a |
| chr13_-_21701323 | 0.55 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr9_-_710896 | 0.54 |
ENSDART00000180478
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr20_+_9683994 | 0.53 |
ENSDART00000053831
|
ptger2b
|
prostaglandin E receptor 2b (subtype EP2) |
| chr14_-_976912 | 0.53 |
ENSDART00000114053
|
arsia
|
arylsulfatase family, member Ia |
| chr22_+_19405517 | 0.52 |
ENSDART00000138245
ENSDART00000155144 |
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr9_+_2499627 | 0.51 |
ENSDART00000160782
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr7_+_14005111 | 0.50 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chrM_+_8894 | 0.50 |
ENSDART00000093611
|
mt-atp8
|
ATP synthase 8, mitochondrial |
| chr13_-_37649595 | 0.49 |
ENSDART00000115354
|
si:dkey-188i13.10
|
si:dkey-188i13.10 |
| chr8_+_48966165 | 0.49 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr10_+_45128375 | 0.47 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr18_+_44849809 | 0.46 |
ENSDART00000097328
|
arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr5_+_24287927 | 0.46 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr24_+_2962988 | 0.46 |
ENSDART00000164449
|
eci2
|
enoyl-CoA delta isomerase 2 |
| chr13_+_30421472 | 0.45 |
ENSDART00000143569
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr19_+_7001170 | 0.44 |
ENSDART00000110366
|
zbtb22b
|
zinc finger and BTB domain containing 22b |
| chr7_+_23875269 | 0.42 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr6_-_43028896 | 0.42 |
ENSDART00000149977
|
glyctk
|
glycerate kinase |
| chr9_-_30346279 | 0.42 |
ENSDART00000089488
|
sytl5
|
synaptotagmin-like 5 |
| chr14_-_1538600 | 0.41 |
ENSDART00000180925
|
CABZ01109480.1
|
|
| chr21_-_18824434 | 0.41 |
ENSDART00000156333
|
si:dkey-112m2.1
|
si:dkey-112m2.1 |
| chr18_+_16750080 | 0.41 |
ENSDART00000136320
|
rnf141
|
ring finger protein 141 |
| chr11_+_37251825 | 0.40 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr4_-_77506362 | 0.40 |
ENSDART00000174387
ENSDART00000181181 |
CABZ01087415.1
|
|
| chr6_+_41957280 | 0.40 |
ENSDART00000050114
|
oxtr
|
oxytocin receptor |
| chr24_+_41940299 | 0.40 |
ENSDART00000022349
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr20_+_23960525 | 0.40 |
ENSDART00000042123
|
cx52.6
|
connexin 52.6 |
| chr15_-_13254480 | 0.40 |
ENSDART00000190499
|
zgc:172282
|
zgc:172282 |
| chr21_+_22845317 | 0.39 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr1_-_22861348 | 0.39 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr1_-_58913813 | 0.38 |
ENSDART00000056494
|
zgc:171687
|
zgc:171687 |
| chr5_-_63644938 | 0.38 |
ENSDART00000050865
|
surf4l
|
surfeit gene 4, like |
| chr21_-_37733287 | 0.38 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr18_+_40993369 | 0.38 |
ENSDART00000141162
|
si:dkey-283j8.1
|
si:dkey-283j8.1 |
| chr22_-_21476836 | 0.37 |
ENSDART00000135977
|
s1pr3a
|
sphingosine-1-phosphate receptor 3a |
| chr5_-_20123002 | 0.37 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr3_-_39648021 | 0.36 |
ENSDART00000055171
|
grapa
|
GRB2-related adaptor protein a |
| chr1_+_46598502 | 0.35 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr19_-_42573219 | 0.35 |
ENSDART00000126021
ENSDART00000133695 ENSDART00000131558 |
zgc:103438
|
zgc:103438 |
| chr12_+_29173523 | 0.35 |
ENSDART00000153212
|
adgra1a
|
adhesion G protein-coupled receptor A1a |
| chr18_-_19095141 | 0.35 |
ENSDART00000090962
ENSDART00000090922 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr15_-_21877726 | 0.35 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr17_+_33496067 | 0.34 |
ENSDART00000184724
|
pth2
|
parathyroid hormone 2 |
| chr16_-_40373836 | 0.34 |
ENSDART00000134498
|
si:dkey-242e21.3
|
si:dkey-242e21.3 |
| chr13_+_7575563 | 0.33 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr21_-_2248239 | 0.33 |
ENSDART00000169448
|
si:ch73-299h12.1
|
si:ch73-299h12.1 |
| chr23_-_11128601 | 0.33 |
ENSDART00000131232
|
cntn3a.2
|
contactin 3a, tandem duplicate 2 |
| chr4_+_7822773 | 0.32 |
ENSDART00000171391
|
si:ch1073-67j19.2
|
si:ch1073-67j19.2 |
| chr18_-_41160975 | 0.30 |
ENSDART00000187517
|
CABZ01005876.1
|
|
| chr4_-_57356512 | 0.30 |
ENSDART00000164760
|
znf1069
|
zinc finger protein 1069 |
| chr7_+_34549377 | 0.30 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr14_+_24934736 | 0.29 |
ENSDART00000191821
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr13_-_33134611 | 0.28 |
ENSDART00000026280
|
plek2
|
pleckstrin 2 |
| chr17_-_5583345 | 0.28 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr14_+_20156477 | 0.28 |
ENSDART00000123434
|
fmr1
|
fragile X mental retardation 1 |
| chr17_+_33495194 | 0.27 |
ENSDART00000033691
|
pth2
|
parathyroid hormone 2 |
| chr18_-_41160579 | 0.27 |
ENSDART00000181480
|
CABZ01005876.1
|
|
| chr14_+_30774515 | 0.27 |
ENSDART00000191666
|
atl3
|
atlastin 3 |
| chr2_-_15362923 | 0.26 |
ENSDART00000166926
|
olfm3b
|
olfactomedin 3b |
| chr18_+_40993196 | 0.26 |
ENSDART00000115111
|
si:dkey-283j8.1
|
si:dkey-283j8.1 |
| chr22_+_17399124 | 0.25 |
ENSDART00000145769
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr13_+_22964868 | 0.25 |
ENSDART00000142129
|
tacr2
|
tachykinin receptor 2 |
| chr2_-_51087077 | 0.24 |
ENSDART00000167987
|
ftr67
|
finTRIM family, member 67 |
| chr15_+_22867174 | 0.24 |
ENSDART00000035812
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr9_+_32358514 | 0.24 |
ENSDART00000144608
|
plcl1
|
phospholipase C like 1 |
| chr4_+_67050047 | 0.23 |
ENSDART00000170257
|
znf1070
|
zinc finger protein 1070 |
| chr11_+_24251141 | 0.23 |
ENSDART00000182684
|
pnp4a
|
purine nucleoside phosphorylase 4a |
| chr17_-_12216465 | 0.22 |
ENSDART00000154056
|
si:rp71-84d9.1
|
si:rp71-84d9.1 |
| chr4_+_41789497 | 0.22 |
ENSDART00000126634
|
si:dkey-237m9.2
|
si:dkey-237m9.2 |
| chr20_-_47270519 | 0.21 |
ENSDART00000153329
|
si:dkeyp-104f11.8
|
si:dkeyp-104f11.8 |
| chr17_+_23556764 | 0.20 |
ENSDART00000146787
|
pank1a
|
pantothenate kinase 1a |
| chr17_+_12942021 | 0.20 |
ENSDART00000192514
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr19_-_10881486 | 0.19 |
ENSDART00000168852
ENSDART00000160438 |
PSMD4 (1 of many)
psmd4a
|
proteasome 26S subunit, non-ATPase 4 proteasome 26S subunit, non-ATPase 4a |
| chr6_+_42475730 | 0.19 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr24_-_38192003 | 0.18 |
ENSDART00000109975
|
crp7
|
C-reactive protein 7 |
| chr2_-_48356787 | 0.18 |
ENSDART00000098004
|
per2
|
period circadian clock 2 |
| chr11_-_6001845 | 0.16 |
ENSDART00000108628
|
ano8b
|
anoctamin 8b |
Network of associatons between targets according to the STRING database.
First level regulatory network of jdp2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.6 | 1.9 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.6 | 1.8 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.4 | 3.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.3 | 1.4 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.3 | 2.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.2 | 1.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.2 | 0.7 | GO:0032263 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.2 | 1.6 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 3.7 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.2 | 0.5 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 5.5 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.1 | 0.6 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 1.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 1.7 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.1 | 0.5 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.4 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 2.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.8 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.7 | GO:0045907 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) positive regulation of vasoconstriction(GO:0045907) |
| 0.1 | 0.7 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 1.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.8 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.3 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.1 | 0.8 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 1.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 1.5 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 1.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.3 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.4 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.7 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 1.0 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.5 | GO:0060971 | peptide hormone processing(GO:0016486) embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.6 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.6 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 1.1 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.0 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 1.2 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 1.0 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.6 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.8 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 1.5 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.9 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 2.6 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.5 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 1.3 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.7 | GO:0021549 | cerebellum development(GO:0021549) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.6 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.1 | 0.3 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 5.5 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.7 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 3.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.8 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 3.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0099503 | secretory vesicle(GO:0099503) |
| 0.0 | 1.0 | GO:0005604 | basement membrane(GO:0005604) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.5 | 3.3 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.4 | 3.7 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.4 | 3.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 5.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 2.4 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.3 | 2.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 0.7 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.2 | 1.1 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.2 | 0.8 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 2.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.0 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.4 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.1 | 0.4 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.1 | 1.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.6 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 1.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.3 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 1.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 1.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 1.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 5.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 3.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.8 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 2.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.0 | 2.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.0 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 4.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 2.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |