Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
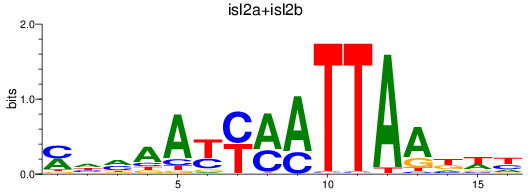
Results for isl2a+isl2b
Z-value: 0.49
Transcription factors associated with isl2a+isl2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
isl2a
|
ENSDARG00000003971 | ISL LIM homeobox 2a |
|
isl2b
|
ENSDARG00000053499 | ISL LIM homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| isl2a | dr11_v1_chr25_-_32751982_32751982 | -0.65 | 2.7e-03 | Click! |
| isl2b | dr11_v1_chr7_+_29992889_29992889 | -0.25 | 3.0e-01 | Click! |
Activity profile of isl2a+isl2b motif
Sorted Z-values of isl2a+isl2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_24002503 | 1.95 |
ENSDART00000164702
|
chia.2
|
chitinase, acidic.2 |
| chr11_+_24001993 | 1.75 |
ENSDART00000168215
|
chia.2
|
chitinase, acidic.2 |
| chr19_-_5125663 | 1.74 |
ENSDART00000005547
ENSDART00000132642 |
gnb3b
|
guanine nucleotide binding protein (G protein), beta polypeptide 3b |
| chr6_-_12588044 | 1.54 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr9_+_2574122 | 1.53 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr2_-_293793 | 1.41 |
ENSDART00000082091
|
si:ch73-40a17.3
|
si:ch73-40a17.3 |
| chr2_-_293036 | 1.29 |
ENSDART00000171629
|
si:ch73-40a17.3
|
si:ch73-40a17.3 |
| chr16_-_28658341 | 1.08 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr6_+_54538948 | 1.07 |
ENSDART00000149270
|
tulp1b
|
tubby like protein 1b |
| chr21_+_11385031 | 1.01 |
ENSDART00000040598
|
cel.2
|
carboxyl ester lipase, tandem duplicate 2 |
| chr10_-_7756865 | 0.99 |
ENSDART00000114373
ENSDART00000125407 ENSDART00000016317 |
loxa
|
lysyl oxidase a |
| chr12_-_4781801 | 0.92 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr23_-_3511630 | 0.92 |
ENSDART00000019667
|
rnf114
|
ring finger protein 114 |
| chr6_+_39905021 | 0.85 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr17_-_200316 | 0.84 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr1_-_9641845 | 0.82 |
ENSDART00000121490
ENSDART00000159411 |
ugt5b2
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B2 UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr6_+_9175886 | 0.82 |
ENSDART00000165333
|
si:ch211-207l14.1
|
si:ch211-207l14.1 |
| chr5_-_41494831 | 0.78 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr12_+_16168342 | 0.77 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr3_-_59297532 | 0.70 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr14_-_4682114 | 0.69 |
ENSDART00000014454
|
gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr17_+_16564921 | 0.67 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr13_-_49666615 | 0.66 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr7_+_24573721 | 0.63 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr25_+_3994823 | 0.60 |
ENSDART00000154020
|
eps8l2
|
EPS8 like 2 |
| chr5_+_69747417 | 0.58 |
ENSDART00000153717
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr9_-_36924388 | 0.56 |
ENSDART00000059756
|
ralba
|
v-ral simian leukemia viral oncogene homolog Ba (ras related) |
| chr7_+_60079302 | 0.55 |
ENSDART00000051524
|
etnppl
|
ethanolamine-phosphate phospho-lyase |
| chr1_-_50859053 | 0.55 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr21_-_20939488 | 0.54 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr8_+_52637507 | 0.54 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr11_-_1400507 | 0.54 |
ENSDART00000173029
ENSDART00000172953 ENSDART00000111140 |
rpl29
|
ribosomal protein L29 |
| chr25_-_27621268 | 0.53 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr8_+_1651821 | 0.53 |
ENSDART00000060865
ENSDART00000186304 |
rasal1b
|
RAS protein activator like 1b (GAP1 like) |
| chr18_+_3579829 | 0.52 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr18_+_924949 | 0.52 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr12_+_24342303 | 0.51 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr14_-_2206476 | 0.50 |
ENSDART00000081870
|
pcdh2ab6
|
protocadherin 2 alpha b 6 |
| chr7_+_22657566 | 0.49 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr23_-_18024543 | 0.49 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr21_-_35325466 | 0.48 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr21_+_45841731 | 0.47 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr20_-_5291012 | 0.47 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr22_-_10470663 | 0.43 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr21_-_44081540 | 0.43 |
ENSDART00000130833
|
FO704810.1
|
|
| chr3_-_25268751 | 0.42 |
ENSDART00000139423
|
mgat3a
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase a |
| chr2_+_47249179 | 0.42 |
ENSDART00000144438
|
si:ch211-284d12.3
|
si:ch211-284d12.3 |
| chr2_-_22927958 | 0.41 |
ENSDART00000141621
|
myo7bb
|
myosin VIIBb |
| chr25_+_35212919 | 0.41 |
ENSDART00000180127
|
ano5a
|
anoctamin 5a |
| chr5_+_22791686 | 0.40 |
ENSDART00000014806
|
npas2
|
neuronal PAS domain protein 2 |
| chr3_-_13068189 | 0.40 |
ENSDART00000167180
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr7_-_71829649 | 0.40 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr12_+_35203091 | 0.40 |
ENSDART00000153022
|
ndst2b
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2b |
| chr4_-_2219705 | 0.39 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr4_-_1801519 | 0.39 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr12_-_6880694 | 0.38 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr11_+_30663300 | 0.37 |
ENSDART00000161662
|
ttbk1a
|
tau tubulin kinase 1a |
| chr20_+_32552912 | 0.36 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr22_-_7168571 | 0.35 |
ENSDART00000165549
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr5_-_19861766 | 0.35 |
ENSDART00000088904
|
gltpa
|
glycolipid transfer protein a |
| chr24_-_6897884 | 0.34 |
ENSDART00000080766
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr10_-_2524297 | 0.34 |
ENSDART00000192475
|
CU856539.1
|
|
| chr25_-_9805269 | 0.34 |
ENSDART00000192048
|
lrrc4c
|
leucine rich repeat containing 4C |
| chr22_-_17844117 | 0.34 |
ENSDART00000159363
|
BX908731.1
|
|
| chr3_+_7808459 | 0.33 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr22_-_14739491 | 0.32 |
ENSDART00000133385
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr17_-_2834764 | 0.32 |
ENSDART00000024027
|
bdkrb1
|
bradykinin receptor B1 |
| chr20_-_47188966 | 0.31 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr23_-_36724575 | 0.31 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr7_-_2047639 | 0.30 |
ENSDART00000173892
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr12_+_10062953 | 0.30 |
ENSDART00000148689
|
slc4a1b
|
solute carrier family 4 (anion exchanger), member 1b (Diego blood group) |
| chr7_-_66868543 | 0.30 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr10_-_43404027 | 0.30 |
ENSDART00000086227
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr24_-_6898302 | 0.29 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr12_+_18681477 | 0.29 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr2_+_49232559 | 0.29 |
ENSDART00000154285
ENSDART00000175147 |
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr20_+_36234335 | 0.29 |
ENSDART00000193484
ENSDART00000181664 |
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr7_-_7692992 | 0.28 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr9_+_54039006 | 0.28 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr25_+_13406069 | 0.28 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr15_+_46356879 | 0.28 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr12_-_4532066 | 0.28 |
ENSDART00000092687
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr19_-_47323267 | 0.28 |
ENSDART00000190077
|
CU138512.1
|
|
| chr19_+_9186175 | 0.27 |
ENSDART00000039325
|
hcn3
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 3 |
| chr20_+_66857 | 0.27 |
ENSDART00000114999
|
lrfn2b
|
leucine rich repeat and fibronectin type III domain containing 2b |
| chr9_+_32069989 | 0.26 |
ENSDART00000139540
|
si:dkey-83m22.7
|
si:dkey-83m22.7 |
| chr14_+_21722235 | 0.26 |
ENSDART00000183667
|
stx3a
|
syntaxin 3A |
| chr15_-_46779934 | 0.25 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr1_-_17803614 | 0.25 |
ENSDART00000138475
|
sorbs2a
|
sorbin and SH3 domain containing 2a |
| chr6_-_52788213 | 0.25 |
ENSDART00000179880
|
rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr18_-_8398972 | 0.24 |
ENSDART00000136512
|
BEND7
|
si:ch211-220f12.4 |
| chr19_-_5769728 | 0.24 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr15_+_9327252 | 0.23 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr19_+_31904836 | 0.23 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr11_-_45138857 | 0.23 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr1_+_55279124 | 0.23 |
ENSDART00000152660
|
si:ch211-286b5.6
|
si:ch211-286b5.6 |
| chr14_+_36521005 | 0.23 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr7_-_50410524 | 0.23 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr9_+_51147380 | 0.22 |
ENSDART00000003913
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr11_-_26832685 | 0.22 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr22_-_17482282 | 0.21 |
ENSDART00000105453
|
si:ch211-197g15.7
|
si:ch211-197g15.7 |
| chr16_+_11660839 | 0.21 |
ENSDART00000193911
ENSDART00000143683 |
si:dkey-250k15.10
|
si:dkey-250k15.10 |
| chr6_-_49510553 | 0.21 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr17_-_40956035 | 0.21 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr13_-_2010191 | 0.21 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr22_-_17482810 | 0.20 |
ENSDART00000171293
|
si:ch211-197g15.7
|
si:ch211-197g15.7 |
| chr11_-_29737088 | 0.20 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr11_+_29856032 | 0.20 |
ENSDART00000079150
|
grpr
|
gastrin-releasing peptide receptor |
| chr9_-_394088 | 0.20 |
ENSDART00000169014
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr12_+_1139690 | 0.19 |
ENSDART00000160442
|
CABZ01072885.1
|
|
| chr15_-_14552101 | 0.19 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr18_+_35128685 | 0.19 |
ENSDART00000151579
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr15_+_25438714 | 0.18 |
ENSDART00000154164
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr16_-_17072440 | 0.18 |
ENSDART00000002493
ENSDART00000178443 |
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr8_+_31716872 | 0.18 |
ENSDART00000161121
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr13_+_38430466 | 0.18 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr25_+_7671640 | 0.18 |
ENSDART00000145367
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr23_-_18707418 | 0.18 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr19_+_24872159 | 0.18 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr7_-_7692723 | 0.18 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr1_+_51721851 | 0.17 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr17_+_15433518 | 0.17 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr1_-_25966068 | 0.17 |
ENSDART00000137869
ENSDART00000134192 |
synpo2b
|
synaptopodin 2b |
| chr18_+_13275735 | 0.17 |
ENSDART00000148127
|
plcg2
|
phospholipase C, gamma 2 |
| chr21_-_35534401 | 0.16 |
ENSDART00000112308
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr8_+_25913787 | 0.16 |
ENSDART00000190257
ENSDART00000062515 |
sema3h
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3H |
| chr11_+_30057762 | 0.16 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr3_-_22216771 | 0.16 |
ENSDART00000130546
|
maptb
|
microtubule-associated protein tau b |
| chr6_+_52350443 | 0.15 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr18_-_3473081 | 0.15 |
ENSDART00000174975
|
myo7aa
|
myosin VIIAa |
| chr10_-_17083180 | 0.14 |
ENSDART00000170083
|
fam166b
|
family with sequence similarity 166, member B |
| chr14_-_24391424 | 0.14 |
ENSDART00000113376
ENSDART00000126894 |
fam13b
|
family with sequence similarity 13, member B |
| chr4_-_5019113 | 0.14 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr1_+_39482588 | 0.14 |
ENSDART00000181790
|
tenm3
|
teneurin transmembrane protein 3 |
| chr21_-_10886709 | 0.14 |
ENSDART00000134408
|
si:dkey-277m11.2
|
si:dkey-277m11.2 |
| chr22_-_10165446 | 0.12 |
ENSDART00000142012
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr4_-_49582758 | 0.12 |
ENSDART00000180834
ENSDART00000187608 |
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr18_+_34225520 | 0.11 |
ENSDART00000126115
|
v2rl1
|
vomeronasal 2 receptor, l1 |
| chr21_-_35419486 | 0.11 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr22_+_19552987 | 0.10 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr3_+_3681116 | 0.10 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr22_+_30137374 | 0.10 |
ENSDART00000187808
|
add3a
|
adducin 3 (gamma) a |
| chr12_-_9516981 | 0.09 |
ENSDART00000106285
|
si:ch211-207i20.3
|
si:ch211-207i20.3 |
| chr13_+_646700 | 0.09 |
ENSDART00000006892
|
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr10_-_2524917 | 0.09 |
ENSDART00000188642
|
CU856539.1
|
|
| chr7_+_66822229 | 0.09 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr4_-_46915962 | 0.09 |
ENSDART00000169555
|
si:ch211-134c10.1
|
si:ch211-134c10.1 |
| chr18_+_43891051 | 0.09 |
ENSDART00000024213
|
cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr7_-_12821277 | 0.09 |
ENSDART00000091584
|
zgc:158785
|
zgc:158785 |
| chr3_+_14388010 | 0.09 |
ENSDART00000171726
ENSDART00000165452 |
tmem56b
|
transmembrane protein 56b |
| chr21_-_2814709 | 0.08 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr16_-_45094599 | 0.08 |
ENSDART00000155479
|
si:rp71-77l1.1
|
si:rp71-77l1.1 |
| chr2_-_1622641 | 0.08 |
ENSDART00000082143
|
prkacbb
|
protein kinase, cAMP-dependent, catalytic, beta b |
| chr20_+_41021054 | 0.08 |
ENSDART00000146052
|
man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr16_+_10972733 | 0.08 |
ENSDART00000049323
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr5_+_55225497 | 0.08 |
ENSDART00000144087
|
tmc2a
|
transmembrane channel-like 2a |
| chr15_+_46357080 | 0.08 |
ENSDART00000155571
ENSDART00000156541 |
wu:fb18f06
|
wu:fb18f06 |
| chr20_+_44082532 | 0.08 |
ENSDART00000188481
|
BX510352.1
|
|
| chr16_+_28932038 | 0.07 |
ENSDART00000149480
|
npr1b
|
natriuretic peptide receptor 1b |
| chr11_-_40165683 | 0.07 |
ENSDART00000186510
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr2_+_20605925 | 0.07 |
ENSDART00000191510
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr11_+_45436703 | 0.07 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr22_-_34995333 | 0.06 |
ENSDART00000110900
|
kcnip2
|
Kv channel interacting protein 2 |
| chr3_-_54846444 | 0.06 |
ENSDART00000074010
|
ubald1b
|
UBA-like domain containing 1b |
| chr13_-_42560662 | 0.06 |
ENSDART00000124898
|
CR792417.1
|
|
| chr6_+_9204099 | 0.05 |
ENSDART00000150167
ENSDART00000115394 |
tbx19
|
T-box 19 |
| chr10_+_31248036 | 0.05 |
ENSDART00000193574
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr22_-_23545307 | 0.05 |
ENSDART00000166915
|
crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
| chr2_+_56657804 | 0.05 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr24_-_26310854 | 0.05 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr21_-_25522510 | 0.05 |
ENSDART00000162711
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr21_-_19314618 | 0.04 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr22_-_23545828 | 0.04 |
ENSDART00000161253
|
crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
| chr10_-_45210184 | 0.03 |
ENSDART00000167128
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr3_-_19200571 | 0.03 |
ENSDART00000131503
ENSDART00000012335 |
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr9_+_12896340 | 0.02 |
ENSDART00000136417
|
si:ch211-167j6.5
|
si:ch211-167j6.5 |
| chr6_-_54486410 | 0.02 |
ENSDART00000064679
|
CR847944.1
|
|
| chr10_-_34916208 | 0.02 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr24_+_33622769 | 0.02 |
ENSDART00000079342
|
dnah5l
|
dynein, axonemal, heavy chain 5 like |
| chr1_-_52210950 | 0.02 |
ENSDART00000083946
|
pld6
|
phospholipase D family, member 6 |
| chr8_-_52229462 | 0.02 |
ENSDART00000185949
ENSDART00000051825 |
tcf7l1b
|
transcription factor 7 like 1b |
| chr11_-_44979281 | 0.02 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr18_-_48547564 | 0.01 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr9_+_24065855 | 0.01 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr24_+_12989727 | 0.01 |
ENSDART00000126842
ENSDART00000129309 |
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr9_+_1039215 | 0.01 |
ENSDART00000192186
|
DOCK9 (1 of many)
|
dedicator of cytokinesis 9 |
| chr25_+_21833287 | 0.01 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr8_+_24747865 | 0.01 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr1_+_55758257 | 0.01 |
ENSDART00000139312
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr14_-_7137808 | 0.01 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr20_+_22799857 | 0.00 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr14_-_4145594 | 0.00 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr23_+_32426954 | 0.00 |
ENSDART00000192529
ENSDART00000172313 |
si:ch211-66i15.5
|
si:ch211-66i15.5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of isl2a+isl2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.2 | 1.5 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.5 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.6 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.3 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.1 | 1.0 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.4 | GO:1901909 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.2 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 1.0 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.3 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:0032608 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.4 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.4 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.7 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.6 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 1.2 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.6 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.5 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.4 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.2 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.4 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.5 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.1 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.2 | GO:0030183 | B cell differentiation(GO:0030183) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.7 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.3 | 1.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.3 | 3.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 1.5 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 0.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 1.7 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.5 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.1 | 0.4 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.5 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 0.5 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.6 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.2 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.1 | 1.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.6 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.7 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 2.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.8 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |