Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
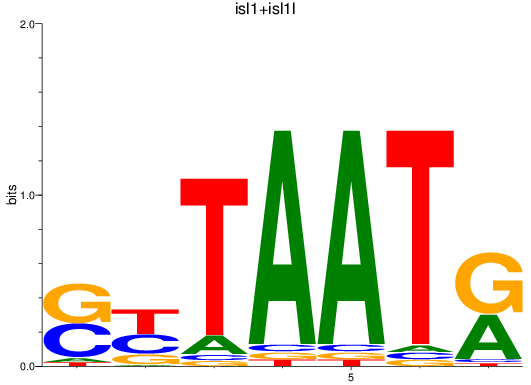
Results for isl1+isl1l
Z-value: 0.77
Transcription factors associated with isl1+isl1l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
isl1
|
ENSDARG00000004023 | ISL LIM homeobox 1 |
|
isl1l
|
ENSDARG00000021055 | islet1, like |
|
isl1l
|
ENSDARG00000110204 | islet1, like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| isl1 | dr11_v1_chr5_-_40734045_40734045 | 0.23 | 3.4e-01 | Click! |
| isl1l | dr11_v1_chr10_+_8680730_8680730 | -0.22 | 3.8e-01 | Click! |
Activity profile of isl1+isl1l motif
Sorted Z-values of isl1+isl1l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_46941930 | 2.20 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr4_+_15968483 | 1.80 |
ENSDART00000101575
|
si:dkey-117n7.5
|
si:dkey-117n7.5 |
| chr14_-_41478265 | 1.39 |
ENSDART00000149886
ENSDART00000016002 |
tspan7
|
tetraspanin 7 |
| chr13_-_50624173 | 1.38 |
ENSDART00000184181
|
vox
|
ventral homeobox |
| chr12_-_13205854 | 1.37 |
ENSDART00000077829
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr14_-_17072736 | 1.33 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr2_+_11685742 | 1.28 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr21_+_723998 | 1.28 |
ENSDART00000160956
|
oaz1b
|
ornithine decarboxylase antizyme 1b |
| chr4_+_5506952 | 1.24 |
ENSDART00000032857
ENSDART00000160222 |
mapk11
|
mitogen-activated protein kinase 11 |
| chr2_-_17114852 | 1.24 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr4_-_5826320 | 1.22 |
ENSDART00000165354
|
foxm1
|
forkhead box M1 |
| chr19_-_47570672 | 1.17 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr22_-_24984733 | 1.09 |
ENSDART00000142147
ENSDART00000187284 |
dnal4b
|
dynein, axonemal, light chain 4b |
| chr7_-_54679595 | 1.08 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr8_+_8936912 | 1.08 |
ENSDART00000135958
|
si:dkey-83k24.5
|
si:dkey-83k24.5 |
| chr13_-_50614639 | 1.05 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr11_-_18107447 | 1.04 |
ENSDART00000187376
|
qrich1
|
glutamine-rich 1 |
| chr11_-_10850936 | 1.04 |
ENSDART00000091901
|
psmd14
|
proteasome 26S subunit, non-ATPase 14 |
| chr24_+_18714212 | 1.04 |
ENSDART00000171181
|
cspp1a
|
centrosome and spindle pole associated protein 1a |
| chr21_-_22317920 | 1.03 |
ENSDART00000191083
ENSDART00000108701 |
gdpd4b
|
glycerophosphodiester phosphodiesterase domain containing 4b |
| chr15_-_23529945 | 1.03 |
ENSDART00000152543
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr2_+_10821579 | 1.01 |
ENSDART00000179694
|
glmna
|
glomulin, FKBP associated protein a |
| chr25_+_30131055 | 1.01 |
ENSDART00000152705
|
api5
|
apoptosis inhibitor 5 |
| chr14_+_16287968 | 1.00 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr5_+_23136544 | 1.00 |
ENSDART00000003428
ENSDART00000109340 ENSDART00000171039 ENSDART00000178821 |
prps1a
|
phosphoribosyl pyrophosphate synthetase 1A |
| chr6_-_18618106 | 0.97 |
ENSDART00000161562
|
znf207b
|
zinc finger protein 207, b |
| chr8_-_1051438 | 0.97 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr11_-_18017287 | 0.96 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr13_+_15701849 | 0.96 |
ENSDART00000003517
|
trmt61a
|
tRNA methyltransferase 61A |
| chr19_+_42432625 | 0.96 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr3_-_23513155 | 0.95 |
ENSDART00000170200
|
BX682558.1
|
|
| chr12_+_27034760 | 0.95 |
ENSDART00000181170
ENSDART00000153054 |
fbrs
|
fibrosin |
| chr6_+_22326624 | 0.93 |
ENSDART00000020333
|
rae1
|
ribonucleic acid export 1 |
| chr5_+_25762271 | 0.92 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr17_+_32343121 | 0.91 |
ENSDART00000156051
|
dhx32b
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32b |
| chr8_+_48942470 | 0.91 |
ENSDART00000005464
ENSDART00000132035 |
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr16_+_53203370 | 0.90 |
ENSDART00000154669
|
si:ch211-269k10.2
|
si:ch211-269k10.2 |
| chr23_+_31596441 | 0.89 |
ENSDART00000053534
|
tbpl1
|
TBP-like 1 |
| chr6_-_53326421 | 0.88 |
ENSDART00000191740
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr2_+_10821127 | 0.87 |
ENSDART00000145770
ENSDART00000174629 ENSDART00000081094 |
glmna
|
glomulin, FKBP associated protein a |
| chr12_-_13205572 | 0.86 |
ENSDART00000152670
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr15_+_36309070 | 0.86 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr7_+_756942 | 0.85 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr22_+_9922301 | 0.84 |
ENSDART00000105924
|
blf
|
bloody fingers |
| chr12_+_14149686 | 0.84 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr20_+_34770197 | 0.84 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr11_-_14813029 | 0.83 |
ENSDART00000004920
ENSDART00000122645 |
sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr4_-_15103646 | 0.83 |
ENSDART00000138183
ENSDART00000181044 |
nrf1
|
nuclear respiratory factor 1 |
| chr11_-_438294 | 0.82 |
ENSDART00000040812
|
nuf2
|
NUF2, NDC80 kinetochore complex component, homolog |
| chr15_-_39969988 | 0.82 |
ENSDART00000146054
|
rps5
|
ribosomal protein S5 |
| chr25_+_36292057 | 0.82 |
ENSDART00000152329
|
bmb
|
brambleberry |
| chr12_-_19028046 | 0.81 |
ENSDART00000142253
ENSDART00000078484 |
rangap1b
|
Ran GTPase activating protein 1b |
| chr4_+_9478500 | 0.81 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr15_-_44052927 | 0.81 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr11_+_6009984 | 0.79 |
ENSDART00000185680
|
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr22_+_438714 | 0.79 |
ENSDART00000136491
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr2_-_25140022 | 0.79 |
ENSDART00000134543
|
nceh1a
|
neutral cholesterol ester hydrolase 1a |
| chr3_-_58650057 | 0.78 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr22_+_9918872 | 0.77 |
ENSDART00000177953
|
blf
|
bloody fingers |
| chr6_+_16031189 | 0.75 |
ENSDART00000015333
|
gbx2
|
gastrulation brain homeobox 2 |
| chr17_-_29271359 | 0.74 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr1_+_51475094 | 0.73 |
ENSDART00000146352
|
meis1a
|
Meis homeobox 1 a |
| chr7_+_36898850 | 0.73 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr7_+_38897836 | 0.72 |
ENSDART00000024330
|
creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr16_-_55259199 | 0.70 |
ENSDART00000161130
|
iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr1_-_19079957 | 0.70 |
ENSDART00000141795
|
phox2ba
|
paired-like homeobox 2ba |
| chr1_+_41588170 | 0.70 |
ENSDART00000139175
|
si:dkey-56e3.2
|
si:dkey-56e3.2 |
| chr7_+_24496894 | 0.70 |
ENSDART00000149994
|
nelfa
|
negative elongation factor complex member A |
| chr7_+_36035432 | 0.69 |
ENSDART00000179004
|
irx3a
|
iroquois homeobox 3a |
| chr12_+_13205955 | 0.69 |
ENSDART00000092906
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr9_-_35633827 | 0.69 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr9_+_14152211 | 0.68 |
ENSDART00000148055
|
si:ch211-67e16.11
|
si:ch211-67e16.11 |
| chr17_-_19535328 | 0.68 |
ENSDART00000077809
|
cyp26c1
|
cytochrome P450, family 26, subfamily C, polypeptide 1 |
| chr7_-_18598661 | 0.67 |
ENSDART00000182109
|
DTX4
|
si:ch211-119e14.2 |
| chr22_-_14115292 | 0.67 |
ENSDART00000105717
ENSDART00000165670 |
aox5
|
aldehyde oxidase 5 |
| chr22_-_621888 | 0.66 |
ENSDART00000135829
|
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr5_-_36597612 | 0.65 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr11_+_6010177 | 0.64 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr7_-_30174882 | 0.64 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr3_-_58116314 | 0.63 |
ENSDART00000154901
|
si:ch211-256e16.6
|
si:ch211-256e16.6 |
| chr20_+_25712276 | 0.62 |
ENSDART00000121585
ENSDART00000185772 |
cep135
|
centrosomal protein 135 |
| chr1_+_59328030 | 0.62 |
ENSDART00000172464
|
CABZ01052576.1
|
|
| chr15_+_888704 | 0.62 |
ENSDART00000182796
|
si:dkey-7i4.9
|
si:dkey-7i4.9 |
| chr16_-_35415004 | 0.62 |
ENSDART00000170401
ENSDART00000157832 ENSDART00000170048 |
taf12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr16_+_9762261 | 0.61 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr7_+_15871156 | 0.61 |
ENSDART00000145946
|
pax6b
|
paired box 6b |
| chr8_-_25033681 | 0.61 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr10_+_7593185 | 0.60 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr10_-_1961576 | 0.60 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr10_+_32104305 | 0.59 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr20_+_25711718 | 0.58 |
ENSDART00000033436
|
cep135
|
centrosomal protein 135 |
| chr22_-_26852516 | 0.57 |
ENSDART00000005829
|
gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr19_-_30420602 | 0.56 |
ENSDART00000144121
|
ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr15_-_25518084 | 0.56 |
ENSDART00000158594
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr21_+_2506013 | 0.55 |
ENSDART00000162351
|
hmgcrb
|
3-hydroxy-3-methylglutaryl-CoA reductase b |
| chr7_-_71585065 | 0.55 |
ENSDART00000128678
|
mettl4
|
methyltransferase like 4 |
| chr7_+_26998169 | 0.55 |
ENSDART00000128110
ENSDART00000101018 |
caprin1a
|
cell cycle associated protein 1a |
| chr5_-_68495224 | 0.55 |
ENSDART00000187955
|
ephb4a
|
eph receptor B4a |
| chr10_-_11353101 | 0.54 |
ENSDART00000092110
|
lin54
|
lin-54 DREAM MuvB core complex component |
| chr15_-_43625549 | 0.54 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr22_-_28644517 | 0.53 |
ENSDART00000189538
|
jagn1b
|
jagunal homolog 1b |
| chr21_+_26522571 | 0.53 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr20_-_22464250 | 0.53 |
ENSDART00000165904
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr6_+_15268685 | 0.53 |
ENSDART00000128090
ENSDART00000154417 |
ecrg4b
|
esophageal cancer related gene 4b |
| chr9_-_14137295 | 0.53 |
ENSDART00000127640
ENSDART00000189018 ENSDART00000188985 |
ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr1_-_43727012 | 0.52 |
ENSDART00000181064
|
bdh2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr6_+_41452979 | 0.52 |
ENSDART00000007353
|
wdr82
|
WD repeat domain 82 |
| chr4_-_12930086 | 0.51 |
ENSDART00000013604
|
lemd3
|
LEM domain containing 3 |
| chr3_-_40162843 | 0.51 |
ENSDART00000129664
ENSDART00000025285 |
drg2
|
developmentally regulated GTP binding protein 2 |
| chr10_-_17587832 | 0.51 |
ENSDART00000113101
|
smarcad1b
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 b |
| chr7_+_15871408 | 0.51 |
ENSDART00000014572
|
pax6b
|
paired box 6b |
| chr1_+_33647682 | 0.50 |
ENSDART00000114384
|
znf654
|
zinc finger protein 654 |
| chr7_+_67429185 | 0.50 |
ENSDART00000162553
ENSDART00000178646 |
kars
|
lysyl-tRNA synthetase |
| chr13_-_42536642 | 0.50 |
ENSDART00000134533
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr18_-_12295092 | 0.50 |
ENSDART00000033248
|
fam107b
|
family with sequence similarity 107, member B |
| chr23_-_16734009 | 0.49 |
ENSDART00000125449
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr13_-_48388726 | 0.49 |
ENSDART00000169473
|
fbxo11a
|
F-box protein 11a |
| chr2_-_10821053 | 0.49 |
ENSDART00000056034
|
rpap2
|
RNA polymerase II associated protein 2 |
| chr4_+_15899668 | 0.49 |
ENSDART00000101592
|
col7a1l
|
collagen type VII alpha 1-like |
| chr14_+_11457500 | 0.48 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr11_+_2172335 | 0.48 |
ENSDART00000170593
|
hoxc12b
|
homeobox C12b |
| chr5_-_42083363 | 0.48 |
ENSDART00000162596
|
cxcl11.5
|
chemokine (C-X-C motif) ligand 11, duplicate 5 |
| chr16_-_36979592 | 0.47 |
ENSDART00000168443
|
snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr6_+_16406723 | 0.47 |
ENSDART00000040035
|
ccdc80l1
|
coiled-coil domain containing 80 like 1 |
| chr12_-_7253270 | 0.46 |
ENSDART00000035762
|
ube2d1b
|
ubiquitin-conjugating enzyme E2D 1b |
| chr2_+_27010439 | 0.46 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr9_+_41459759 | 0.46 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr1_-_35924495 | 0.45 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr8_-_53926228 | 0.45 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr15_-_24588649 | 0.45 |
ENSDART00000060469
|
gemin4
|
gem (nuclear organelle) associated protein 4 |
| chr11_+_24316110 | 0.45 |
ENSDART00000186028
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr9_+_3283608 | 0.45 |
ENSDART00000192275
|
hat1
|
histone acetyltransferase 1 |
| chr5_-_56948058 | 0.45 |
ENSDART00000083074
ENSDART00000191028 |
si:ch211-127d4.3
|
si:ch211-127d4.3 |
| chr17_-_33716688 | 0.44 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr25_+_7532627 | 0.44 |
ENSDART00000187660
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr22_+_16308806 | 0.44 |
ENSDART00000162685
|
lrrc39
|
leucine rich repeat containing 39 |
| chr11_+_30647545 | 0.44 |
ENSDART00000114792
|
gb:eh507706
|
expressed sequence EH507706 |
| chr6_-_40581376 | 0.44 |
ENSDART00000185412
|
tspo
|
translocator protein |
| chr12_+_11356592 | 0.44 |
ENSDART00000148770
|
si:rp71-19m20.1
|
si:rp71-19m20.1 |
| chr17_-_36529016 | 0.44 |
ENSDART00000025019
|
colec11
|
collectin sub-family member 11 |
| chr9_-_27720612 | 0.43 |
ENSDART00000000566
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr16_+_9400661 | 0.43 |
ENSDART00000146174
|
ice1
|
KIAA0947-like (H. sapiens) |
| chr13_+_48359573 | 0.43 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr13_-_36391496 | 0.43 |
ENSDART00000100217
ENSDART00000140243 |
actn1
|
actinin, alpha 1 |
| chr21_-_21178410 | 0.42 |
ENSDART00000185277
ENSDART00000141341 ENSDART00000145872 ENSDART00000079678 |
ftsj1
|
FtsJ RNA methyltransferase homolog 1 |
| chr7_-_34192834 | 0.42 |
ENSDART00000125131
|
smad6a
|
SMAD family member 6a |
| chr2_+_1881334 | 0.42 |
ENSDART00000161420
|
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr8_+_18830759 | 0.41 |
ENSDART00000089079
|
mpnd
|
MPN domain containing |
| chr4_+_17539134 | 0.41 |
ENSDART00000013313
|
itfg2
|
integrin alpha FG-GAP repeat containing 2 |
| chr3_-_18737126 | 0.41 |
ENSDART00000055767
|
e4f1
|
E4F transcription factor 1 |
| chr25_+_36348401 | 0.41 |
ENSDART00000103006
|
hist1h2a3
|
histone cluster 1 H2A family member 3 |
| chr20_-_5400395 | 0.40 |
ENSDART00000168103
|
snw1
|
SNW domain containing 1 |
| chr20_-_38787341 | 0.40 |
ENSDART00000136771
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr12_+_27034594 | 0.40 |
ENSDART00000111679
|
fbrs
|
fibrosin |
| chr20_+_18703951 | 0.40 |
ENSDART00000191070
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr12_+_26706745 | 0.40 |
ENSDART00000141401
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr23_+_20431140 | 0.40 |
ENSDART00000193950
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr25_-_27541621 | 0.40 |
ENSDART00000130678
|
spam1
|
sperm adhesion molecule 1 |
| chr13_-_9159852 | 0.40 |
ENSDART00000170185
ENSDART00000158697 ENSDART00000143393 ENSDART00000164973 ENSDART00000159910 |
HTRA2 (1 of many)
|
si:dkey-112g5.16 |
| chr22_-_9934011 | 0.39 |
ENSDART00000168223
|
si:dkey-253d23.9
|
si:dkey-253d23.9 |
| chr22_-_20309283 | 0.39 |
ENSDART00000182125
ENSDART00000048775 |
si:dkey-110c1.10
|
si:dkey-110c1.10 |
| chr23_+_20431388 | 0.39 |
ENSDART00000132920
ENSDART00000102963 ENSDART00000109899 ENSDART00000140219 |
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr6_-_31682135 | 0.39 |
ENSDART00000153988
|
cachd1
|
cache domain containing 1 |
| chr20_+_39686193 | 0.38 |
ENSDART00000153142
|
hddc2
|
HD domain containing 2 |
| chr1_+_30946231 | 0.38 |
ENSDART00000022841
|
metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr3_+_13860815 | 0.37 |
ENSDART00000162451
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr17_+_21760032 | 0.37 |
ENSDART00000190425
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr10_-_2943474 | 0.37 |
ENSDART00000188698
|
oclna
|
occludin a |
| chr20_+_29209926 | 0.37 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr16_-_29194517 | 0.37 |
ENSDART00000046114
ENSDART00000148899 |
mef2d
|
myocyte enhancer factor 2d |
| chr13_-_36582341 | 0.36 |
ENSDART00000137335
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr3_+_24062748 | 0.36 |
ENSDART00000188574
|
cbx1a
|
chromobox homolog 1a (HP1 beta homolog Drosophila) |
| chr23_+_26142807 | 0.36 |
ENSDART00000158878
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr4_+_59711338 | 0.35 |
ENSDART00000150849
|
si:dkey-149m13.4
|
si:dkey-149m13.4 |
| chr12_-_3299149 | 0.35 |
ENSDART00000182032
|
lsm12b
|
LSM12 homolog b |
| chr14_+_1240419 | 0.35 |
ENSDART00000181248
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr24_-_32665283 | 0.35 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr18_-_50839033 | 0.35 |
ENSDART00000169773
|
ddb1
|
damage-specific DNA binding protein 1 |
| chr6_+_36844127 | 0.35 |
ENSDART00000020505
|
chd1l
|
chromodomain helicase DNA binding protein 1-like |
| chr8_-_51367298 | 0.35 |
ENSDART00000060628
|
chmp7
|
charged multivesicular body protein 7 |
| chr12_+_28910762 | 0.35 |
ENSDART00000076342
ENSDART00000160939 ENSDART00000076572 |
rnf40
|
ring finger protein 40 |
| chr21_-_7781555 | 0.35 |
ENSDART00000084380
ENSDART00000189131 |
aggf1
|
angiogenic factor with G patch and FHA domains 1 |
| chr5_-_64203101 | 0.34 |
ENSDART00000029364
|
ak5l
|
adenylate kinase 5, like |
| chr3_-_21062706 | 0.34 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr25_+_19489370 | 0.34 |
ENSDART00000091132
|
glsl
|
glutaminase like |
| chr2_-_7189594 | 0.34 |
ENSDART00000139703
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr14_-_31693148 | 0.34 |
ENSDART00000172958
|
map7d3
|
MAP7 domain containing 3 |
| chr23_+_36653376 | 0.34 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr5_+_59449762 | 0.33 |
ENSDART00000150230
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr24_-_1023112 | 0.33 |
ENSDART00000147508
ENSDART00000066863 |
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr4_+_29909870 | 0.33 |
ENSDART00000150324
|
znf1111
|
zinc finger protein 1111 |
| chr7_-_38664947 | 0.32 |
ENSDART00000100546
ENSDART00000112405 |
c6ast1
|
six-cysteine containing astacin protease 1 |
| chr4_+_52356485 | 0.32 |
ENSDART00000170639
|
zgc:173705
|
zgc:173705 |
| chr9_-_41025062 | 0.32 |
ENSDART00000002053
|
pms1
|
PMS1 homolog 1, mismatch repair system component |
| chr21_+_31253048 | 0.32 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr7_+_8543317 | 0.31 |
ENSDART00000114998
|
jac8
|
jacalin 8 |
| chr21_-_30408775 | 0.31 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr20_-_38787047 | 0.31 |
ENSDART00000152913
ENSDART00000153430 |
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr18_+_35861930 | 0.30 |
ENSDART00000185223
|
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr11_-_16395956 | 0.30 |
ENSDART00000115085
|
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of isl1+isl1l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.6 | 2.2 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.3 | 1.3 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.3 | 0.8 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.3 | 0.8 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.2 | 1.0 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.2 | 1.0 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 1.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.2 | 2.4 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.2 | 1.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 0.6 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.2 | 0.2 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.2 | 1.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 0.5 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.2 | 0.5 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 0.5 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.2 | 0.9 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 | 1.0 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.2 | 0.8 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.2 | 0.5 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 1.1 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.2 | 0.5 | GO:0042373 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.4 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.1 | 0.4 | GO:0006290 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.9 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 0.9 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.8 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.1 | 1.8 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.8 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.7 | GO:0055014 | outflow tract morphogenesis(GO:0003151) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 1.2 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.1 | 1.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.5 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 1.2 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 1.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.4 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.1 | 0.6 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.4 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.9 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.3 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 1.3 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.1 | 0.7 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.1 | 1.2 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.3 | GO:0006591 | arginine biosynthetic process(GO:0006526) ornithine metabolic process(GO:0006591) |
| 0.1 | 0.5 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.1 | 0.4 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.9 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.3 | GO:0019884 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 1.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.4 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 0.7 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.8 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.5 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.7 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.4 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.4 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.2 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.5 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.5 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.4 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.6 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0044821 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.4 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.7 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.1 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.5 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.2 | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) |
| 0.0 | 0.1 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.6 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.4 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.5 | GO:0071219 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.7 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 1.2 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 0.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.9 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0072380 | TRC complex(GO:0072380) |
| 0.2 | 1.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 1.0 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 0.6 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 0.4 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.8 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 1.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.3 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.9 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 1.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 1.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.4 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.0 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.9 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.0 | GO:0044279 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.3 | 1.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 1.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 1.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 0.7 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.2 | 1.0 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.2 | 1.2 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.4 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.4 | GO:0032405 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.5 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.7 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 0.2 | GO:0034246 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.1 | 0.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.4 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.6 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.5 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 1.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.9 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.6 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.0 | 0.4 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.6 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.5 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.4 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.4 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 1.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.0 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 1.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.4 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 2.2 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 0.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0016405 | CoA-ligase activity(GO:0016405) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 3.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.6 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.9 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |