Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
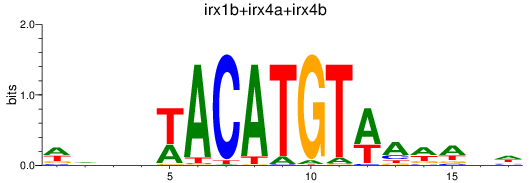
Results for irx1b+irx4a+irx4b
Z-value: 0.24
Transcription factors associated with irx1b+irx4a+irx4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irx4a
|
ENSDARG00000035648 | iroquois homeobox 4a |
|
irx4b
|
ENSDARG00000036051 | iroquois homeobox 4b |
|
irx1b
|
ENSDARG00000056594 | iroquois homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irx4a | dr11_v1_chr16_-_383664_383664 | -0.89 | 4.0e-07 | Click! |
| irx4b | dr11_v1_chr19_+_28115801_28115801 | -0.50 | 2.9e-02 | Click! |
| irx1b | dr11_v1_chr19_-_28130658_28130658 | 0.43 | 6.8e-02 | Click! |
Activity profile of irx1b+irx4a+irx4b motif
Sorted Z-values of irx1b+irx4a+irx4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_-_29072162 | 0.67 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr7_-_18877109 | 0.47 |
ENSDART00000113593
|
mllt3
|
MLLT3, super elongation complex subunit |
| chr18_+_22109379 | 0.46 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr15_-_23522653 | 0.41 |
ENSDART00000144685
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr17_-_5352924 | 0.41 |
ENSDART00000167275
|
supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr7_+_56577906 | 0.34 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr5_-_58996324 | 0.33 |
ENSDART00000033923
|
mis12
|
MIS12 kinetochore complex component |
| chr7_+_56577522 | 0.28 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr14_-_10617127 | 0.28 |
ENSDART00000154299
|
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr10_-_22912255 | 0.26 |
ENSDART00000131992
|
si:ch1073-143l10.2
|
si:ch1073-143l10.2 |
| chr21_+_20549395 | 0.25 |
ENSDART00000181633
|
efna5a
|
ephrin-A5a |
| chr10_-_45002196 | 0.24 |
ENSDART00000170767
|
ved
|
ventrally expressed dharma/bozozok antagonist |
| chr16_-_44709832 | 0.22 |
ENSDART00000156784
|
si:ch211-151m7.6
|
si:ch211-151m7.6 |
| chr14_-_26392146 | 0.20 |
ENSDART00000037999
|
b4galt7
|
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) |
| chr8_+_26410197 | 0.18 |
ENSDART00000145836
ENSDART00000053447 |
ifrd2
|
interferon-related developmental regulator 2 |
| chr20_+_23440632 | 0.18 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr1_+_53919110 | 0.18 |
ENSDART00000020680
|
nup133
|
nucleoporin 133 |
| chr21_-_45871866 | 0.17 |
ENSDART00000161716
|
larp1
|
La ribonucleoprotein domain family, member 1 |
| chr2_-_7185460 | 0.16 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr19_+_3653976 | 0.16 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr6_-_34838397 | 0.14 |
ENSDART00000060169
ENSDART00000169605 |
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr2_-_13333932 | 0.13 |
ENSDART00000150238
ENSDART00000168258 |
si:dkey-185p13.1
vps4b
|
si:dkey-185p13.1 vacuolar protein sorting 4b homolog B (S. cerevisiae) |
| chr20_-_23440955 | 0.13 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr8_+_23784471 | 0.13 |
ENSDART00000189457
|
si:ch211-163l21.8
|
si:ch211-163l21.8 |
| chr5_+_66170479 | 0.10 |
ENSDART00000172117
|
gldc
|
glycine dehydrogenase (decarboxylating) |
| chr24_-_25098719 | 0.09 |
ENSDART00000193651
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr22_+_1940595 | 0.08 |
ENSDART00000163506
|
znf1167
|
zinc finger protein 1167 |
| chr23_+_19701587 | 0.08 |
ENSDART00000104425
|
dnase1l1
|
deoxyribonuclease I-like 1 |
| chr5_-_35159379 | 0.08 |
ENSDART00000144105
|
fcho2
|
FCH domain only 2 |
| chr3_-_402714 | 0.07 |
ENSDART00000134062
ENSDART00000105659 |
mhc1zja
|
major histocompatibility complex class I ZJA |
| chr22_+_16320076 | 0.06 |
ENSDART00000164161
|
osbpl1a
|
oxysterol binding protein-like 1A |
| chr17_-_14705039 | 0.05 |
ENSDART00000154281
ENSDART00000123550 |
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr8_+_29986265 | 0.05 |
ENSDART00000148258
|
ptch1
|
patched 1 |
| chr21_+_12036238 | 0.04 |
ENSDART00000102463
ENSDART00000155426 |
zgc:162344
|
zgc:162344 |
| chr7_-_13362590 | 0.03 |
ENSDART00000091616
|
sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr10_-_28117740 | 0.03 |
ENSDART00000134491
|
med13a
|
mediator complex subunit 13a |
| chr8_-_23783633 | 0.02 |
ENSDART00000132657
|
si:ch211-163l21.7
|
si:ch211-163l21.7 |
| chr5_-_41103583 | 0.02 |
ENSDART00000051070
ENSDART00000074781 |
golph3
|
golgi phosphoprotein 3 |
| chr21_+_43882274 | 0.02 |
ENSDART00000075672
|
sra1
|
steroid receptor RNA activator 1 |
| chr18_+_5549672 | 0.01 |
ENSDART00000184970
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr4_-_5691257 | 0.01 |
ENSDART00000110497
|
tmem63a
|
transmembrane protein 63A |
| chr15_-_41734639 | 0.01 |
ENSDART00000154230
ENSDART00000167443 |
ftr90
|
finTRIM family, member 90 |
| chr16_-_7828838 | 0.00 |
ENSDART00000191434
ENSDART00000108653 |
tcaim
|
T cell activation inhibitor, mitochondrial |
| chr5_+_34578479 | 0.00 |
ENSDART00000016314
|
gfm2
|
G elongation factor, mitochondrial 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irx1b+irx4a+irx4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 0.3 | GO:0044034 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.2 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.1 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.3 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |