Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
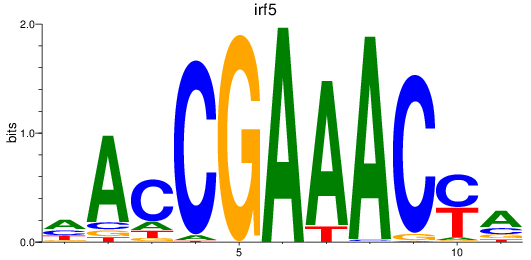
Results for irf5
Z-value: 0.70
Transcription factors associated with irf5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf5
|
ENSDARG00000045681 | interferon regulatory factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf5 | dr11_v1_chr4_-_13613148_13613148 | 0.63 | 3.7e-03 | Click! |
Activity profile of irf5 motif
Sorted Z-values of irf5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_36127234 | 1.33 |
ENSDART00000130917
|
coil
|
coilin p80 |
| chr19_+_14059349 | 1.31 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr18_-_16922905 | 1.29 |
ENSDART00000187165
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr25_+_27410352 | 1.26 |
ENSDART00000154362
|
pot1
|
protection of telomeres 1 homolog |
| chr22_+_336256 | 1.20 |
ENSDART00000019155
|
btg2
|
B-cell translocation gene 2 |
| chr25_+_16646113 | 1.18 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr16_-_32303835 | 1.17 |
ENSDART00000191408
|
mms22l
|
MMS22-like, DNA repair protein |
| chr24_-_37688398 | 1.15 |
ENSDART00000141414
|
zgc:112185
|
zgc:112185 |
| chr22_-_16291041 | 1.08 |
ENSDART00000021666
|
rtca
|
RNA 3'-terminal phosphate cyclase |
| chr9_-_39968820 | 1.07 |
ENSDART00000100311
|
IKZF2
|
si:zfos-1425h8.1 |
| chr8_+_23802384 | 1.05 |
ENSDART00000137820
|
si:ch211-163l21.8
|
si:ch211-163l21.8 |
| chr15_-_33818872 | 1.04 |
ENSDART00000158325
|
n4bp2l2
|
NEDD4 binding protein 2-like 2 |
| chr13_+_24263049 | 1.04 |
ENSDART00000135992
ENSDART00000088005 |
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr6_-_33916756 | 1.02 |
ENSDART00000137447
ENSDART00000138488 |
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr10_-_22150419 | 1.01 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr9_+_32178374 | 1.00 |
ENSDART00000078576
|
coq10b
|
coenzyme Q10B |
| chr19_+_10832092 | 0.98 |
ENSDART00000191851
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr21_+_20903244 | 0.98 |
ENSDART00000186193
|
c7b
|
complement component 7b |
| chr14_+_15768942 | 0.97 |
ENSDART00000158998
|
ergic1
|
endoplasmic reticulum-golgi intermediate compartment 1 |
| chr2_-_24554416 | 0.97 |
ENSDART00000052061
|
cnn2
|
calponin 2 |
| chr22_+_18319230 | 0.96 |
ENSDART00000184747
ENSDART00000184649 |
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr23_+_27652906 | 0.95 |
ENSDART00000077931
|
wnt1
|
wingless-type MMTV integration site family, member 1 |
| chr5_+_57442271 | 0.94 |
ENSDART00000097395
|
prpf4
|
PRP4 pre-mRNA processing factor 4 homolog (yeast) |
| chr3_+_21200763 | 0.94 |
ENSDART00000067841
|
zgc:112038
|
zgc:112038 |
| chr7_+_69187585 | 0.94 |
ENSDART00000160499
ENSDART00000166258 |
marveld3
|
MARVEL domain containing 3 |
| chr1_+_38153944 | 0.94 |
ENSDART00000135666
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr20_+_1316495 | 0.93 |
ENSDART00000064439
|
nup43
|
nucleoporin 43 |
| chr9_+_23003208 | 0.93 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr12_-_16452200 | 0.92 |
ENSDART00000037601
|
rpp30
|
ribonuclease P/MRP 30 subunit |
| chr13_-_33700461 | 0.92 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr12_+_23424108 | 0.92 |
ENSDART00000077732
|
bambia
|
BMP and activin membrane-bound inhibitor (Xenopus laevis) homolog a |
| chr23_+_31942040 | 0.90 |
ENSDART00000088607
|
nemp1
|
nuclear envelope integral membrane protein 1 |
| chr20_+_36806398 | 0.88 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr17_+_12658411 | 0.88 |
ENSDART00000139918
|
gpn1
|
GPN-loop GTPase 1 |
| chr5_+_44805269 | 0.87 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr9_-_30264415 | 0.87 |
ENSDART00000060150
|
mid1ip1a
|
MID1 interacting protein 1a |
| chr3_-_4501026 | 0.86 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr21_+_19319804 | 0.86 |
ENSDART00000063621
|
abraxas2a
|
abraxas 2a, BRISC complex subunit |
| chr1_-_35924495 | 0.84 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr15_+_25452092 | 0.81 |
ENSDART00000009545
|
pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr6_-_9646275 | 0.79 |
ENSDART00000012903
|
wdr12
|
WD repeat domain 12 |
| chr2_+_38025260 | 0.78 |
ENSDART00000075905
|
hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr20_+_1316803 | 0.78 |
ENSDART00000152586
ENSDART00000152165 |
nup43
|
nucleoporin 43 |
| chr5_-_69716501 | 0.78 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr8_+_31008287 | 0.78 |
ENSDART00000129882
ENSDART00000180694 |
ptgesl
|
prostaglandin E synthase 2-like |
| chr20_-_43743700 | 0.77 |
ENSDART00000100620
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr4_+_12931763 | 0.77 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr13_-_23051766 | 0.76 |
ENSDART00000111774
|
supv3l1
|
SUV3-like helicase |
| chr15_-_36369743 | 0.73 |
ENSDART00000155942
|
si:dkey-23k10.3
|
si:dkey-23k10.3 |
| chr9_+_890536 | 0.73 |
ENSDART00000139132
|
steap3
|
STEAP family member 3, metalloreductase |
| chr19_+_4968947 | 0.72 |
ENSDART00000003634
ENSDART00000134808 |
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr18_+_16133595 | 0.72 |
ENSDART00000080423
|
ctsd
|
cathepsin D |
| chr21_+_33187992 | 0.72 |
ENSDART00000162745
ENSDART00000188388 |
BX072577.1
|
|
| chr5_+_58455488 | 0.72 |
ENSDART00000038602
ENSDART00000127958 |
slc37a2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr17_-_12966907 | 0.72 |
ENSDART00000022874
|
psma6a
|
proteasome subunit alpha 6a |
| chr6_-_10728057 | 0.71 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr25_+_28862660 | 0.71 |
ENSDART00000154681
|
si:ch211-106e7.2
|
si:ch211-106e7.2 |
| chr16_+_48616220 | 0.71 |
ENSDART00000004751
ENSDART00000130045 |
pbx2
|
pre-B-cell leukemia homeobox 2 |
| chr24_-_34680956 | 0.71 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr14_-_41467497 | 0.70 |
ENSDART00000181220
|
mid1ip1l
|
MID1 interacting protein 1, like |
| chr5_+_41793001 | 0.69 |
ENSDART00000136439
ENSDART00000190477 ENSDART00000192289 |
bcl7a
|
BCL tumor suppressor 7A |
| chr10_-_31563049 | 0.67 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr19_-_29303788 | 0.67 |
ENSDART00000112167
|
srfbp1
|
serum response factor binding protein 1 |
| chr4_-_11580948 | 0.67 |
ENSDART00000049066
|
net1
|
neuroepithelial cell transforming 1 |
| chr22_+_18319666 | 0.67 |
ENSDART00000033103
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr2_-_59157790 | 0.67 |
ENSDART00000192303
ENSDART00000159362 |
ftr32
|
finTRIM family, member 32 |
| chr7_+_32693890 | 0.66 |
ENSDART00000121972
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr4_-_17669881 | 0.66 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr5_+_57726425 | 0.65 |
ENSDART00000134684
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr12_+_17154655 | 0.65 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr24_-_3407507 | 0.65 |
ENSDART00000132648
|
nck1b
|
NCK adaptor protein 1b |
| chr10_+_35152928 | 0.65 |
ENSDART00000063418
|
nsun5
|
NOP2/Sun domain family, member 5 |
| chr6_-_23002373 | 0.65 |
ENSDART00000037709
ENSDART00000170369 |
nol11
|
nucleolar protein 11 |
| chr19_+_42227400 | 0.64 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr14_+_22076596 | 0.63 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr1_-_55750208 | 0.63 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr6_-_45869127 | 0.63 |
ENSDART00000062459
ENSDART00000180563 |
rbm19
|
RNA binding motif protein 19 |
| chr15_-_1484795 | 0.63 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr11_-_6206520 | 0.62 |
ENSDART00000150199
ENSDART00000148246 ENSDART00000019440 |
pole4
|
polymerase (DNA-directed), epsilon 4, accessory subunit |
| chr16_+_46666074 | 0.62 |
ENSDART00000074919
ENSDART00000142698 |
ubqln4
|
ubiquilin 4 |
| chr7_+_38808027 | 0.62 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr19_+_20163826 | 0.61 |
ENSDART00000090942
ENSDART00000134650 |
ccdc126
|
coiled-coil domain containing 126 |
| chr7_+_46020508 | 0.61 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr7_+_49681040 | 0.60 |
ENSDART00000176372
ENSDART00000192172 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr23_-_27152866 | 0.59 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr8_+_17168114 | 0.58 |
ENSDART00000183901
|
cenph
|
centromere protein H |
| chr7_-_20865005 | 0.57 |
ENSDART00000190752
|
fis1
|
fission, mitochondrial 1 |
| chr21_+_35215810 | 0.57 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr16_+_19637384 | 0.57 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr9_+_41459759 | 0.57 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr23_+_42346799 | 0.56 |
ENSDART00000159985
ENSDART00000172144 |
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr25_+_8447565 | 0.56 |
ENSDART00000142090
|
fanci
|
Fanconi anemia, complementation group I |
| chr1_-_49530615 | 0.56 |
ENSDART00000113315
|
si:ch211-281g13.5
|
si:ch211-281g13.5 |
| chr6_+_54078703 | 0.56 |
ENSDART00000110845
|
fhit
|
fragile histidine triad gene |
| chr19_-_10214264 | 0.56 |
ENSDART00000053300
ENSDART00000148225 |
znf865
|
zinc finger protein 865 |
| chr14_-_16151055 | 0.55 |
ENSDART00000162431
|
ptcd3
|
pentatricopeptide repeat domain 3 |
| chr7_+_19482877 | 0.55 |
ENSDART00000077868
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr20_-_23171430 | 0.55 |
ENSDART00000109234
|
spata18
|
spermatogenesis associated 18 |
| chr23_-_29553430 | 0.54 |
ENSDART00000157773
ENSDART00000126384 |
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr16_+_4654333 | 0.53 |
ENSDART00000167665
|
LIN28A
|
si:ch1073-284b18.2 |
| chr5_+_44804791 | 0.53 |
ENSDART00000122288
|
ctsla
|
cathepsin La |
| chr10_+_7719796 | 0.53 |
ENSDART00000191795
|
ggcx
|
gamma-glutamyl carboxylase |
| chr16_+_42772678 | 0.53 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr22_-_6499055 | 0.53 |
ENSDART00000142235
ENSDART00000183588 |
si:dkey-19a16.7
|
si:dkey-19a16.7 |
| chr5_+_69716458 | 0.52 |
ENSDART00000159594
|
mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr12_+_16452575 | 0.52 |
ENSDART00000058665
|
kif20bb
|
kinesin family member 20Bb |
| chr12_+_16452912 | 0.52 |
ENSDART00000192467
|
kif20bb
|
kinesin family member 20Bb |
| chr12_-_33805366 | 0.52 |
ENSDART00000030566
|
galk1
|
galactokinase 1 |
| chr24_+_19578935 | 0.51 |
ENSDART00000137175
|
sulf1
|
sulfatase 1 |
| chr5_+_44805028 | 0.51 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr3_+_25990052 | 0.50 |
ENSDART00000007398
|
gcat
|
glycine C-acetyltransferase |
| chr5_+_36895860 | 0.50 |
ENSDART00000134493
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr5_+_24128908 | 0.49 |
ENSDART00000029719
|
si:ch211-114c12.2
|
si:ch211-114c12.2 |
| chr7_+_8670398 | 0.49 |
ENSDART00000164195
|
si:ch211-1o7.2
|
si:ch211-1o7.2 |
| chr3_+_25999477 | 0.48 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr17_+_46739693 | 0.48 |
ENSDART00000097810
|
pimr22
|
Pim proto-oncogene, serine/threonine kinase, related 22 |
| chr23_-_29553691 | 0.48 |
ENSDART00000053804
|
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr5_-_29531948 | 0.47 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr15_+_1534644 | 0.47 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr22_-_4439311 | 0.47 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr12_+_24060894 | 0.46 |
ENSDART00000021298
|
asb3
|
ankyrin repeat and SOCS box containing 3 |
| chr15_-_2640966 | 0.45 |
ENSDART00000063320
|
cldne
|
claudin e |
| chr7_+_19615056 | 0.45 |
ENSDART00000124752
ENSDART00000190297 |
si:ch211-212k18.15
|
si:ch211-212k18.15 |
| chr19_-_42462491 | 0.45 |
ENSDART00000131715
|
psmb4
|
proteasome subunit beta 4 |
| chr22_-_17499513 | 0.45 |
ENSDART00000105460
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr8_+_45361775 | 0.45 |
ENSDART00000015193
|
chmp4bb
|
charged multivesicular body protein 4Bb |
| chr9_+_23808546 | 0.44 |
ENSDART00000041992
|
dhrs12
|
dehydrogenase/reductase (SDR family) member 12 |
| chr21_-_22122312 | 0.44 |
ENSDART00000101726
|
slc35f2
|
solute carrier family 35, member F2 |
| chr15_+_31911989 | 0.44 |
ENSDART00000111472
|
brca2
|
breast cancer 2, early onset |
| chr10_+_9410304 | 0.44 |
ENSDART00000080843
|
ndufa8
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 |
| chr9_+_24708653 | 0.43 |
ENSDART00000065136
|
CT030712.1
|
|
| chr3_-_55650417 | 0.43 |
ENSDART00000171441
|
axin2
|
axin 2 (conductin, axil) |
| chr21_-_30273418 | 0.43 |
ENSDART00000187069
ENSDART00000181492 |
zgc:175066
|
zgc:175066 |
| chr11_+_10909183 | 0.43 |
ENSDART00000064860
|
rbms1a
|
RNA binding motif, single stranded interacting protein 1a |
| chr12_-_3077395 | 0.43 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr2_+_58377395 | 0.42 |
ENSDART00000193511
|
vapal
|
VAMP (vesicle-associated membrane protein)-associated protein A, like |
| chr19_+_26922780 | 0.42 |
ENSDART00000187396
ENSDART00000188978 |
nelfe
|
negative elongation factor complex member E |
| chr18_-_34170918 | 0.42 |
ENSDART00000015079
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr18_-_21640389 | 0.41 |
ENSDART00000100857
|
slc38a8a
|
solute carrier family 38, member 8a |
| chr7_+_19483277 | 0.41 |
ENSDART00000173750
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr13_+_25364324 | 0.41 |
ENSDART00000187471
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr23_+_44349252 | 0.41 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr4_-_28375127 | 0.40 |
ENSDART00000190198
|
tprkb
|
Tp53rk binding protein |
| chr20_-_1120056 | 0.40 |
ENSDART00000040680
|
rngtt
|
RNA guanylyltransferase and 5'-phosphatase |
| chr7_+_54222156 | 0.40 |
ENSDART00000165201
ENSDART00000158518 |
pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr2_-_21438492 | 0.39 |
ENSDART00000046098
|
plcd1b
|
phospholipase C, delta 1b |
| chr12_+_35650321 | 0.39 |
ENSDART00000190446
|
BX255898.1
|
|
| chr9_+_29040425 | 0.39 |
ENSDART00000150201
|
MRAS
|
si:ch73-116o1.2 |
| chr22_-_30973791 | 0.39 |
ENSDART00000104728
|
ssuh2.2
|
ssu-2 homolog, tandem duplicate 2 |
| chr25_+_20272145 | 0.39 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr6_-_53334259 | 0.39 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr5_+_66326004 | 0.39 |
ENSDART00000144351
|
malt1
|
MALT paracaspase 1 |
| chr18_+_44532668 | 0.38 |
ENSDART00000140672
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr1_-_7570181 | 0.38 |
ENSDART00000103588
|
mxa
|
myxovirus (influenza) resistance A |
| chr8_+_44475793 | 0.37 |
ENSDART00000190118
|
si:ch73-211l2.3
|
si:ch73-211l2.3 |
| chr22_+_10645088 | 0.37 |
ENSDART00000142265
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr6_-_1566581 | 0.37 |
ENSDART00000192993
|
trim107
|
tripartite motif containing 107 |
| chr10_-_31562695 | 0.36 |
ENSDART00000186456
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr3_+_32571929 | 0.36 |
ENSDART00000151025
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr16_-_14552199 | 0.36 |
ENSDART00000133368
|
si:dkey-237j11.3
|
si:dkey-237j11.3 |
| chr23_+_642395 | 0.35 |
ENSDART00000186995
|
irf10
|
interferon regulatory factor 10 |
| chr3_-_55650771 | 0.35 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr7_-_6444011 | 0.35 |
ENSDART00000173010
|
zgc:112234
|
zgc:112234 |
| chr18_+_17624295 | 0.35 |
ENSDART00000151865
ENSDART00000190408 ENSDART00000189204 ENSDART00000188894 ENSDART00000033762 ENSDART00000193471 |
nlrc5
|
NLR family, CARD domain containing 5 |
| chr12_-_28349026 | 0.35 |
ENSDART00000183768
ENSDART00000152998 |
zgc:195081
|
zgc:195081 |
| chr13_+_39324865 | 0.35 |
ENSDART00000115386
|
BX324188.1
|
|
| chr4_-_20108833 | 0.35 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr10_+_2742499 | 0.34 |
ENSDART00000122847
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr9_-_41818760 | 0.33 |
ENSDART00000140601
|
si:dkeyp-30e7.2
|
si:dkeyp-30e7.2 |
| chr5_+_39099172 | 0.33 |
ENSDART00000006079
|
bmp2k
|
BMP2 inducible kinase |
| chr17_-_2039511 | 0.33 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr2_+_34967022 | 0.33 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr11_-_20071642 | 0.32 |
ENSDART00000162931
ENSDART00000159928 ENSDART00000191443 ENSDART00000121722 |
si:dkey-274m17.3
|
si:dkey-274m17.3 |
| chr20_+_6535438 | 0.32 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr13_+_2523032 | 0.32 |
ENSDART00000172261
|
lhb
|
luteinizing hormone, beta polypeptide |
| chr17_-_12408109 | 0.32 |
ENSDART00000155509
|
ankef1b
|
ankyrin repeat and EF-hand domain containing 1b |
| chr14_-_33277743 | 0.32 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr23_+_642001 | 0.32 |
ENSDART00000030643
ENSDART00000124850 |
irf10
|
interferon regulatory factor 10 |
| chr20_+_26683933 | 0.31 |
ENSDART00000139852
ENSDART00000077751 |
foxq1b
|
forkhead box Q1b |
| chr10_-_9410068 | 0.31 |
ENSDART00000041382
|
morn5
|
MORN repeat containing 5 |
| chr15_+_17321218 | 0.31 |
ENSDART00000143796
|
cltcb
|
clathrin, heavy chain b (Hc) |
| chr2_-_6115688 | 0.30 |
ENSDART00000081663
|
prdx1
|
peroxiredoxin 1 |
| chr10_-_15879569 | 0.30 |
ENSDART00000136789
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr16_+_33144306 | 0.30 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr2_+_27403300 | 0.30 |
ENSDART00000099180
|
elovl8a
|
ELOVL fatty acid elongase 8a |
| chr4_-_6373735 | 0.30 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr3_-_29688370 | 0.30 |
ENSDART00000151147
ENSDART00000151679 |
si:ch73-233k15.2
|
si:ch73-233k15.2 |
| chr23_-_45022681 | 0.29 |
ENSDART00000102640
|
ctc1
|
CTS telomere maintenance complex component 1 |
| chr7_+_19600262 | 0.29 |
ENSDART00000007310
|
zgc:171731
|
zgc:171731 |
| chr19_+_9113932 | 0.29 |
ENSDART00000060442
|
setdb1a
|
SET domain, bifurcated 1a |
| chr22_-_17688868 | 0.29 |
ENSDART00000012336
ENSDART00000147070 |
tjp3
|
tight junction protein 3 |
| chr2_+_10007113 | 0.29 |
ENSDART00000155213
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr22_+_32235922 | 0.29 |
ENSDART00000114531
ENSDART00000136125 |
rbm15b
|
RNA binding motif protein 15B |
| chr7_-_19600181 | 0.28 |
ENSDART00000100757
|
oxa1l
|
oxidase (cytochrome c) assembly 1-like |
| chr3_+_24190207 | 0.28 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr16_-_41990421 | 0.28 |
ENSDART00000055921
|
pycard
|
PYD and CARD domain containing |
| chr11_-_3381343 | 0.28 |
ENSDART00000002545
|
mcrs1
|
microspherule protein 1 |
| chr18_+_44532370 | 0.28 |
ENSDART00000086952
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr20_+_26939742 | 0.28 |
ENSDART00000138369
ENSDART00000062061 ENSDART00000152992 |
cdca4
|
cell division cycle associated 4 |
| chr2_+_15586632 | 0.28 |
ENSDART00000164903
|
BX510342.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of irf5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.3 | 0.9 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.3 | 1.3 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.3 | 1.0 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 0.7 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 1.2 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.2 | 0.6 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.2 | 0.7 | GO:0015677 | copper ion import(GO:0015677) |
| 0.2 | 1.0 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.1 | 0.4 | GO:0044878 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 1.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.4 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 1.0 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.1 | 0.8 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.6 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.6 | GO:0015961 | diadenosine polyphosphate metabolic process(GO:0015959) diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.1 | 0.5 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.8 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.4 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.5 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 1.0 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 1.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.5 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.2 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.8 | GO:0007530 | sex determination(GO:0007530) |
| 0.1 | 0.3 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 0.9 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.1 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.3 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 1.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.5 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.9 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.5 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 0.7 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.1 | 0.8 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 1.0 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.1 | 0.2 | GO:0035522 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 1.0 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.8 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.4 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.4 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.9 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.5 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.2 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.9 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 1.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.4 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.3 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 1.0 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 1.6 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.3 | GO:0071451 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.3 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.3 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.9 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.3 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.0 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.0 | 0.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 1.2 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 1.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 2.5 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.6 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.7 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.3 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.7 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.6 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 0.6 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.2 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.3 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 0.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.4 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.6 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.4 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.8 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 1.0 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.4 | GO:0070972 | protein localization to endoplasmic reticulum(GO:0070972) |
| 0.0 | 0.3 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.8 | GO:0002573 | myeloid leukocyte differentiation(GO:0002573) |
| 0.0 | 0.2 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.6 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.5 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.7 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.0 | 0.1 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 0.9 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 0.6 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 1.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 0.6 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.9 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 1.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 1.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.3 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.6 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.3 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.2 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 1.3 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 1.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.4 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.9 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 1.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 1.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.7 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 3.0 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.4 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.1 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.3 | 1.3 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 0.7 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 0.7 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.2 | 1.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.2 | 0.8 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 0.7 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.9 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.5 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 0.5 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.1 | 1.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.5 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.4 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.0 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.5 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 0.2 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.5 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.7 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 1.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.5 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.4 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.8 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 0.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 2.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 0.7 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 2.1 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.8 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |