Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
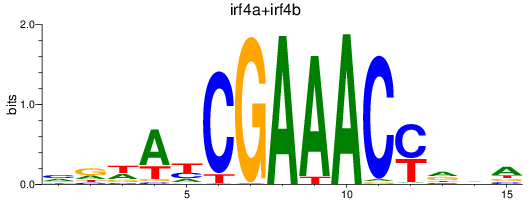
Results for irf4a+irf4b
Z-value: 0.67
Transcription factors associated with irf4a+irf4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf4a
|
ENSDARG00000006560 | interferon regulatory factor 4a |
|
irf4b
|
ENSDARG00000055374 | interferon regulatory factor 4b |
|
irf4b
|
ENSDARG00000115381 | interferon regulatory factor 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf4b | dr11_v1_chr20_+_26556372_26556372 | -0.33 | 1.7e-01 | Click! |
| irf4a | dr11_v1_chr2_-_879800_879800 | 0.03 | 9.1e-01 | Click! |
Activity profile of irf4a+irf4b motif
Sorted Z-values of irf4a+irf4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_68562464 | 0.88 |
ENSDART00000192954
|
BX548011.4
|
|
| chr16_+_46497149 | 0.83 |
ENSDART00000135151
ENSDART00000058324 |
rpz4
|
rapunzel 4 |
| chr9_-_23765480 | 0.80 |
ENSDART00000027212
|
si:ch211-219a4.6
|
si:ch211-219a4.6 |
| chr9_+_23765587 | 0.78 |
ENSDART00000145120
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr15_-_29556757 | 0.75 |
ENSDART00000060049
|
hspa13
|
heat shock protein 70 family, member 13 |
| chr6_+_15762647 | 0.73 |
ENSDART00000127133
ENSDART00000128939 |
iqca1
|
IQ motif containing with AAA domain 1 |
| chr25_+_16646113 | 0.73 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr18_-_24988645 | 0.71 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr12_+_35650321 | 0.70 |
ENSDART00000190446
|
BX255898.1
|
|
| chr20_-_3911546 | 0.68 |
ENSDART00000169787
|
cnksr3
|
cnksr family member 3 |
| chr15_+_31911989 | 0.67 |
ENSDART00000111472
|
brca2
|
breast cancer 2, early onset |
| chr15_+_2421729 | 0.66 |
ENSDART00000082294
ENSDART00000156428 |
hephl1a
|
hephaestin-like 1a |
| chr1_+_31674297 | 0.65 |
ENSDART00000044214
|
wbp1lb
|
WW domain binding protein 1-like b |
| chr2_-_59157790 | 0.59 |
ENSDART00000192303
ENSDART00000159362 |
ftr32
|
finTRIM family, member 32 |
| chr23_-_35396845 | 0.58 |
ENSDART00000142038
ENSDART00000049373 ENSDART00000181978 ENSDART00000171357 |
cmtr1
|
cap methyltransferase 1 |
| chr4_-_12795436 | 0.57 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr15_+_2421432 | 0.57 |
ENSDART00000193772
|
hephl1a
|
hephaestin-like 1a |
| chr20_-_51547464 | 0.51 |
ENSDART00000099486
|
disp1
|
dispatched homolog 1 (Drosophila) |
| chr18_-_23874929 | 0.49 |
ENSDART00000134910
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr25_+_28862660 | 0.48 |
ENSDART00000154681
|
si:ch211-106e7.2
|
si:ch211-106e7.2 |
| chr5_-_13766651 | 0.46 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr13_+_24263049 | 0.45 |
ENSDART00000135992
ENSDART00000088005 |
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr8_-_27687095 | 0.45 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr4_-_12795030 | 0.45 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr15_-_40175894 | 0.43 |
ENSDART00000156632
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr23_-_30045661 | 0.42 |
ENSDART00000122239
ENSDART00000103480 |
ccdc187
|
coiled-coil domain containing 187 |
| chr14_-_25309058 | 0.42 |
ENSDART00000159569
|
htr4
|
5-hydroxytryptamine receptor 4 |
| chr19_+_43669122 | 0.41 |
ENSDART00000139151
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr4_-_17669881 | 0.41 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr15_-_17071328 | 0.39 |
ENSDART00000122617
|
si:ch211-24o10.6
|
si:ch211-24o10.6 |
| chr16_+_27383717 | 0.38 |
ENSDART00000132329
ENSDART00000136256 |
stx17
|
syntaxin 17 |
| chr1_-_1885516 | 0.37 |
ENSDART00000122187
ENSDART00000131675 |
si:ch211-132g1.3
|
si:ch211-132g1.3 |
| chr1_-_35924495 | 0.36 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr17_+_40879331 | 0.36 |
ENSDART00000046003
|
slc4a1ap
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr7_+_19615056 | 0.36 |
ENSDART00000124752
ENSDART00000190297 |
si:ch211-212k18.15
|
si:ch211-212k18.15 |
| chr5_+_28260158 | 0.36 |
ENSDART00000181434
|
ncaph
|
non-SMC condensin I complex, subunit H |
| chr21_+_45626136 | 0.36 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr2_-_43739740 | 0.35 |
ENSDART00000113849
|
kif5ba
|
kinesin family member 5B, a |
| chr1_-_7570181 | 0.35 |
ENSDART00000103588
|
mxa
|
myxovirus (influenza) resistance A |
| chr8_+_38564942 | 0.34 |
ENSDART00000085371
|
rfk
|
riboflavin kinase |
| chr25_+_36045072 | 0.33 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr3_-_49554912 | 0.32 |
ENSDART00000159392
|
CR847534.1
|
|
| chr10_+_7703251 | 0.32 |
ENSDART00000165134
|
ggcx
|
gamma-glutamyl carboxylase |
| chr2_+_15586632 | 0.32 |
ENSDART00000164903
|
BX510342.1
|
|
| chr7_+_42461850 | 0.32 |
ENSDART00000190350
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr18_-_23875219 | 0.32 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr23_-_12906228 | 0.32 |
ENSDART00000138807
|
ndnl2
|
necdin-like 2 |
| chr3_+_15773991 | 0.30 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr12_-_35936329 | 0.30 |
ENSDART00000166634
|
rnf213b
|
ring finger protein 213b |
| chr24_-_26885897 | 0.29 |
ENSDART00000180512
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr19_-_7110617 | 0.29 |
ENSDART00000104838
|
psmb8a
|
proteasome subunit beta 8A |
| chr1_+_38153944 | 0.29 |
ENSDART00000135666
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr13_+_17694845 | 0.28 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr24_-_11467549 | 0.28 |
ENSDART00000082264
|
pxdc1b
|
PX domain containing 1b |
| chr13_+_22476742 | 0.28 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
| chr6_+_41554794 | 0.27 |
ENSDART00000165424
|
srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr23_-_29553430 | 0.27 |
ENSDART00000157773
ENSDART00000126384 |
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr5_+_59397739 | 0.26 |
ENSDART00000148659
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr6_-_18560190 | 0.25 |
ENSDART00000168636
|
tefm
|
transcription elongation factor, mitochondrial |
| chr17_-_22552678 | 0.25 |
ENSDART00000079401
|
si:ch211-125o16.4
|
si:ch211-125o16.4 |
| chr2_+_2818645 | 0.25 |
ENSDART00000163587
|
rock1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr6_-_10728057 | 0.25 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr19_-_791016 | 0.24 |
ENSDART00000037515
|
msto1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr6_-_18400548 | 0.24 |
ENSDART00000179797
ENSDART00000164891 |
trim25
|
tripartite motif containing 25 |
| chr20_-_6476705 | 0.23 |
ENSDART00000077095
|
trappc8
|
trafficking protein particle complex 8 |
| chr11_-_30508843 | 0.23 |
ENSDART00000101667
ENSDART00000179930 |
map4k3a
|
mitogen-activated protein kinase kinase kinase kinase 3a |
| chr15_-_42760110 | 0.23 |
ENSDART00000152490
|
si:ch211-181d7.3
|
si:ch211-181d7.3 |
| chr19_+_791538 | 0.22 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr1_-_36771712 | 0.22 |
ENSDART00000148386
|
prmt9
|
protein arginine methyltransferase 9 |
| chr12_+_36109507 | 0.22 |
ENSDART00000175409
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr22_-_10605045 | 0.22 |
ENSDART00000184812
|
bap1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr18_-_25905574 | 0.22 |
ENSDART00000143899
ENSDART00000163369 |
sema4ba
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ba |
| chr3_-_4501026 | 0.20 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr15_-_1835189 | 0.20 |
ENSDART00000154369
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr16_+_35661331 | 0.19 |
ENSDART00000161725
|
map7d1a
|
MAP7 domain containing 1a |
| chr8_-_51340773 | 0.19 |
ENSDART00000060633
|
kansl3
|
KAT8 regulatory NSL complex subunit 3 |
| chr21_+_17301790 | 0.19 |
ENSDART00000145057
|
tsc1b
|
TSC complex subunit 1b |
| chr8_-_51340931 | 0.19 |
ENSDART00000178209
|
kansl3
|
KAT8 regulatory NSL complex subunit 3 |
| chr19_+_10832092 | 0.19 |
ENSDART00000191851
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr14_-_25309360 | 0.18 |
ENSDART00000088940
|
htr4
|
5-hydroxytryptamine receptor 4 |
| chr22_-_8692305 | 0.17 |
ENSDART00000181602
|
CR450686.4
|
|
| chr23_-_29553691 | 0.17 |
ENSDART00000053804
|
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr12_+_36109071 | 0.17 |
ENSDART00000171268
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr7_-_26579465 | 0.17 |
ENSDART00000173820
|
si:dkey-62k3.6
|
si:dkey-62k3.6 |
| chr3_+_36127287 | 0.17 |
ENSDART00000058605
ENSDART00000182500 |
scpep1
|
serine carboxypeptidase 1 |
| chr4_-_18434924 | 0.16 |
ENSDART00000190271
|
socs2
|
suppressor of cytokine signaling 2 |
| chr15_-_41734639 | 0.16 |
ENSDART00000154230
ENSDART00000167443 |
ftr90
|
finTRIM family, member 90 |
| chr8_-_18229169 | 0.16 |
ENSDART00000131764
ENSDART00000143036 ENSDART00000145986 |
si:ch211-241d21.5
|
si:ch211-241d21.5 |
| chr7_-_19614916 | 0.15 |
ENSDART00000169029
|
zgc:194655
|
zgc:194655 |
| chr20_-_35012093 | 0.15 |
ENSDART00000062761
|
cnstb
|
consortin, connexin sorting protein b |
| chr5_+_65157576 | 0.15 |
ENSDART00000159599
|
cfap157
|
cilia and flagella associated protein 157 |
| chr7_+_19482877 | 0.15 |
ENSDART00000077868
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr22_+_25236657 | 0.15 |
ENSDART00000138012
|
zgc:172218
|
zgc:172218 |
| chr9_+_23003208 | 0.14 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr15_-_43995028 | 0.14 |
ENSDART00000172485
ENSDART00000186320 |
nlrc3l
|
NLR family, CARD domain containing 3-like |
| chr6_+_34511886 | 0.14 |
ENSDART00000179450
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr16_-_41667101 | 0.13 |
ENSDART00000084528
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr4_-_26107841 | 0.13 |
ENSDART00000172012
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr9_-_32837860 | 0.13 |
ENSDART00000142227
|
mxe
|
myxovirus (influenza virus) resistance E |
| chr17_+_35362851 | 0.12 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr21_+_25793970 | 0.12 |
ENSDART00000101217
|
CLDN4 (1 of many)
|
zgc:136892 |
| chr8_+_47897734 | 0.12 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr5_+_21922534 | 0.11 |
ENSDART00000148092
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr20_+_13781779 | 0.11 |
ENSDART00000142999
ENSDART00000152471 |
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr2_-_13333932 | 0.11 |
ENSDART00000150238
ENSDART00000168258 |
si:dkey-185p13.1
vps4b
|
si:dkey-185p13.1 vacuolar protein sorting 4b homolog B (S. cerevisiae) |
| chr1_-_7582859 | 0.11 |
ENSDART00000110696
|
mxb
|
myxovirus (influenza) resistance B |
| chr7_+_19483277 | 0.10 |
ENSDART00000173750
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr3_-_26171668 | 0.10 |
ENSDART00000113890
|
rabep2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr5_+_31959954 | 0.09 |
ENSDART00000142826
|
myo1hb
|
myosin IHb |
| chr21_-_22543611 | 0.09 |
ENSDART00000177084
|
myo5b
|
myosin VB |
| chr17_+_43895158 | 0.09 |
ENSDART00000111123
|
msh4
|
mutS homolog 4 |
| chr9_-_30576522 | 0.08 |
ENSDART00000101085
|
morc3a
|
MORC family CW-type zinc finger 3a |
| chr4_-_8611841 | 0.08 |
ENSDART00000067322
|
fbxl14b
|
F-box and leucine-rich repeat protein 14b |
| chr4_+_11464255 | 0.07 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr12_-_28975705 | 0.06 |
ENSDART00000129173
|
znf646
|
zinc finger protein 646 |
| chr21_-_37933833 | 0.06 |
ENSDART00000184138
ENSDART00000177664 |
FO704750.1
|
|
| chr15_+_34069746 | 0.06 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr4_+_66668652 | 0.04 |
ENSDART00000169500
|
si:ch211-149o24.4
|
si:ch211-149o24.4 |
| chr6_-_52235118 | 0.04 |
ENSDART00000191243
|
tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr2_-_24554416 | 0.03 |
ENSDART00000052061
|
cnn2
|
calponin 2 |
| chr12_-_23658888 | 0.03 |
ENSDART00000088319
|
map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr5_+_66063807 | 0.03 |
ENSDART00000122405
|
jak2b
|
Janus kinase 2b |
| chr17_+_30448452 | 0.03 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr7_+_17908235 | 0.03 |
ENSDART00000077113
|
mta2
|
metastasis associated 1 family, member 2 |
| chr7_-_20464133 | 0.03 |
ENSDART00000078192
|
cnpy4
|
canopy4 |
| chr25_+_15997957 | 0.03 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr18_-_23875370 | 0.02 |
ENSDART00000130163
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr6_+_11250316 | 0.02 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr20_+_2227860 | 0.02 |
ENSDART00000152321
|
tmem200a
|
transmembrane protein 200A |
| chr10_+_8101729 | 0.02 |
ENSDART00000138875
ENSDART00000123447 ENSDART00000185333 |
pstpip2
|
proline-serine-threonine phosphatase interacting protein 2 |
| chr9_+_42269059 | 0.02 |
ENSDART00000113435
|
si:dkey-10c21.1
|
si:dkey-10c21.1 |
| chr6_-_1566581 | 0.02 |
ENSDART00000192993
|
trim107
|
tripartite motif containing 107 |
| chr22_+_19365220 | 0.02 |
ENSDART00000132781
ENSDART00000135672 ENSDART00000153630 |
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr2_-_37140423 | 0.02 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr25_-_483808 | 0.01 |
ENSDART00000104720
|
si:ch1073-385f13.3
|
si:ch1073-385f13.3 |
| chr9_+_34089156 | 0.01 |
ENSDART00000000005
|
ccdc80
|
coiled-coil domain containing 80 |
| chr17_-_40879108 | 0.01 |
ENSDART00000010362
|
supt7l
|
SPT7-like STAGA complex gamma subunit |
| chr20_+_38525567 | 0.00 |
ENSDART00000147787
|
znf512
|
zinc finger protein 512 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irf4a+irf4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0060843 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.1 | 0.6 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.3 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 0.5 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.3 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.2 | GO:0039535 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.1 | 1.0 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 | 0.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.3 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.4 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.1 | 0.4 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.8 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.7 | GO:0008585 | sex determination(GO:0007530) female gonad development(GO:0008585) |
| 0.0 | 0.3 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.6 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.4 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.1 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.5 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.1 | GO:0014005 | microglia development(GO:0014005) |
| 0.0 | 0.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.2 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 0.7 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.4 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 0.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 0.6 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.6 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |