Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
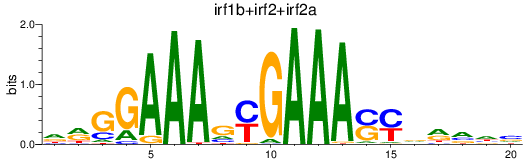
Results for irf1b+irf2+irf2a
Z-value: 1.67
Transcription factors associated with irf1b+irf2+irf2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf2a
|
ENSDARG00000007387 | interferon regulatory factor 2a |
|
irf2
|
ENSDARG00000040465 | interferon regulatory factor 2 |
|
irf1b
|
ENSDARG00000043249 | interferon regulatory factor 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf2a | dr11_v1_chr1_+_39865748_39865753 | 0.57 | 1.1e-02 | Click! |
| irf1b | dr11_v1_chr21_+_45626136_45626136 | 0.54 | 1.6e-02 | Click! |
| irf2 | dr11_v1_chr14_-_4120636_4120636 | 0.21 | 3.9e-01 | Click! |
Activity profile of irf1b+irf2+irf2a motif
Sorted Z-values of irf1b+irf2+irf2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_30387930 | 3.32 |
ENSDART00000112827
|
si:dkey-18p12.4
|
si:dkey-18p12.4 |
| chr8_-_40555340 | 2.72 |
ENSDART00000163348
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr18_-_38270430 | 2.16 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr1_+_53377432 | 1.98 |
ENSDART00000177581
|
ucp1
|
uncoupling protein 1 |
| chr24_+_12945803 | 1.97 |
ENSDART00000005105
|
psme1
|
proteasome activator subunit 1 |
| chr11_+_43043171 | 1.91 |
ENSDART00000180344
|
CABZ01092982.1
|
|
| chr9_+_1138323 | 1.87 |
ENSDART00000190352
ENSDART00000190387 |
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr11_-_472547 | 1.78 |
ENSDART00000005923
|
zgc:77375
|
zgc:77375 |
| chr1_-_47089818 | 1.78 |
ENSDART00000132378
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr11_-_23332592 | 1.78 |
ENSDART00000125024
|
golt1a
|
golgi transport 1A |
| chr21_-_40557281 | 1.77 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr18_+_26422124 | 1.66 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr19_-_7115229 | 1.62 |
ENSDART00000001930
|
psmb13a
|
proteasome subunit beta 13a |
| chr22_+_5687615 | 1.59 |
ENSDART00000133241
ENSDART00000019854 ENSDART00000138102 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr11_-_20096018 | 1.58 |
ENSDART00000030420
|
ogfrl2
|
opioid growth factor receptor-like 2 |
| chr17_-_15149192 | 1.58 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr10_+_17776981 | 1.57 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr9_-_44939104 | 1.57 |
ENSDART00000192903
|
vil1
|
villin 1 |
| chr19_+_7115223 | 1.54 |
ENSDART00000001359
|
psmb12
|
proteasome subunit beta 12 |
| chr21_-_22648007 | 1.51 |
ENSDART00000121788
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr17_-_29192987 | 1.50 |
ENSDART00000164302
|
sptbn5
|
spectrin, beta, non-erythrocytic 5 |
| chr22_+_29990448 | 1.48 |
ENSDART00000165313
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr13_-_3351708 | 1.46 |
ENSDART00000042875
|
si:ch73-193i2.2
|
si:ch73-193i2.2 |
| chr12_+_35650321 | 1.44 |
ENSDART00000190446
|
BX255898.1
|
|
| chr22_+_19290199 | 1.44 |
ENSDART00000148173
|
si:dkey-21e2.15
|
si:dkey-21e2.15 |
| chr9_-_43001898 | 1.38 |
ENSDART00000138515
|
ttn.2
|
titin, tandem duplicate 2 |
| chr5_+_4338874 | 1.36 |
ENSDART00000141866
|
sat1a.1
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 1 |
| chr13_-_37631092 | 1.33 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr22_+_19289970 | 1.30 |
ENSDART00000137976
ENSDART00000132386 |
si:dkey-21e2.15
|
si:dkey-21e2.15 |
| chr10_-_22095505 | 1.29 |
ENSDART00000140210
|
ponzr10
|
plac8 onzin related protein 10 |
| chr24_+_15277216 | 1.28 |
ENSDART00000081449
|
socs6a
|
suppressor of cytokine signaling 6a |
| chr13_+_13681681 | 1.28 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr15_-_17868870 | 1.27 |
ENSDART00000170950
|
atf5b
|
activating transcription factor 5b |
| chr20_+_49787584 | 1.26 |
ENSDART00000193458
ENSDART00000181511 ENSDART00000185850 ENSDART00000185613 ENSDART00000191671 |
CABZ01078261.1
|
|
| chr9_-_18215644 | 1.26 |
ENSDART00000145876
|
epsti1
|
epithelial stromal interaction 1 |
| chr4_-_240737 | 1.25 |
ENSDART00000166186
|
RERG
|
si:cabz01085950.1 |
| chr11_-_45385803 | 1.23 |
ENSDART00000173329
|
trappc10
|
trafficking protein particle complex 10 |
| chr11_-_2250767 | 1.22 |
ENSDART00000018131
|
hnrnpa1a
|
heterogeneous nuclear ribonucleoprotein A1a |
| chr17_-_23609210 | 1.22 |
ENSDART00000064003
|
ifit15
|
interferon-induced protein with tetratricopeptide repeats 15 |
| chr4_+_70782916 | 1.20 |
ENSDART00000187960
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr5_+_36614196 | 1.19 |
ENSDART00000150574
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr1_-_30510839 | 1.19 |
ENSDART00000168189
ENSDART00000174868 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr21_+_5494561 | 1.17 |
ENSDART00000169728
|
ly6m3
|
lymphocyte antigen 6 family member M3 |
| chr1_-_7930679 | 1.17 |
ENSDART00000146090
|
si:dkey-79f11.10
|
si:dkey-79f11.10 |
| chr5_-_12701603 | 1.08 |
ENSDART00000091124
|
aifm3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr16_-_46587938 | 1.07 |
ENSDART00000181433
|
BX323793.1
|
|
| chr20_+_32481348 | 1.07 |
ENSDART00000185018
|
ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr15_-_15227541 | 1.06 |
ENSDART00000184787
|
rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr14_-_2933185 | 1.05 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr20_+_34605044 | 1.05 |
ENSDART00000153376
|
si:ch211-242b18.1
|
si:ch211-242b18.1 |
| chr15_-_44077937 | 1.05 |
ENSDART00000110112
|
lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr1_-_8141135 | 1.05 |
ENSDART00000152295
|
FAM83G
|
si:dkeyp-9d4.4 |
| chr20_-_4793450 | 1.05 |
ENSDART00000053870
|
galca
|
galactosylceramidase a |
| chr9_-_27442339 | 1.04 |
ENSDART00000138602
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr21_+_20901505 | 1.03 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr25_-_12804450 | 1.03 |
ENSDART00000169717
|
ca5a
|
carbonic anhydrase Va |
| chr23_+_45229198 | 1.02 |
ENSDART00000172445
|
ttc39b
|
tetratricopeptide repeat domain 39B |
| chr25_+_35189555 | 1.02 |
ENSDART00000044453
|
ano5a
|
anoctamin 5a |
| chr2_-_42035250 | 1.02 |
ENSDART00000056460
ENSDART00000140788 |
gbp1
|
guanylate binding protein 1 |
| chr1_-_56948694 | 1.01 |
ENSDART00000152597
|
si:ch211-1f22.1
|
si:ch211-1f22.1 |
| chr20_+_44582318 | 1.00 |
ENSDART00000149000
ENSDART00000149775 ENSDART00000085416 |
atad2b
|
ATPase family, AAA domain containing 2B |
| chr6_-_57539141 | 0.99 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr8_+_50983551 | 0.97 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr23_-_21763598 | 0.97 |
ENSDART00000145408
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr6_+_59029485 | 0.97 |
ENSDART00000050140
|
CABZ01088367.1
|
|
| chr3_-_53508580 | 0.97 |
ENSDART00000073978
|
zgc:171711
|
zgc:171711 |
| chr1_-_52201266 | 0.97 |
ENSDART00000143805
ENSDART00000023757 |
rab3da
|
RAB3D, member RAS oncogene family, a |
| chr7_-_40656148 | 0.96 |
ENSDART00000142315
|
nom1
|
nucleolar protein with MIF4G domain 1 |
| chr1_+_38858399 | 0.96 |
ENSDART00000165454
|
CU915762.1
|
|
| chr1_-_47161996 | 0.96 |
ENSDART00000053153
|
mhc1zba
|
major histocompatibility complex class I ZBA |
| chr6_-_45869127 | 0.96 |
ENSDART00000062459
ENSDART00000180563 |
rbm19
|
RNA binding motif protein 19 |
| chr25_-_12803723 | 0.95 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr17_-_23416897 | 0.95 |
ENSDART00000163391
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr23_-_37575030 | 0.95 |
ENSDART00000031875
|
tor1l3
|
torsin family 1 like 3 |
| chr15_-_14083028 | 0.94 |
ENSDART00000147796
ENSDART00000043492 ENSDART00000133080 |
trappc6bl
|
trafficking protein particle complex 6b-like |
| chr11_+_1584747 | 0.93 |
ENSDART00000154583
|
si:dkey-40c23.2
|
si:dkey-40c23.2 |
| chr9_+_42269059 | 0.93 |
ENSDART00000113435
|
si:dkey-10c21.1
|
si:dkey-10c21.1 |
| chr22_-_5518117 | 0.92 |
ENSDART00000164613
|
CABZ01064972.1
|
|
| chr16_-_2650341 | 0.92 |
ENSDART00000128169
ENSDART00000155432 ENSDART00000103722 |
lyplal1
|
lysophospholipase-like 1 |
| chr15_-_43995028 | 0.92 |
ENSDART00000172485
ENSDART00000186320 |
nlrc3l
|
NLR family, CARD domain containing 3-like |
| chr12_+_17436904 | 0.91 |
ENSDART00000079130
|
atad1b
|
ATPase family, AAA domain containing 1b |
| chr8_-_27687095 | 0.90 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr25_-_10503043 | 0.90 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr6_+_38427357 | 0.89 |
ENSDART00000148678
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr3_+_832788 | 0.89 |
ENSDART00000193025
|
FP102888.2
|
|
| chr9_-_14273652 | 0.89 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr9_-_19762801 | 0.89 |
ENSDART00000146841
|
si:ch211-141e20.2
|
si:ch211-141e20.2 |
| chr4_-_31993240 | 0.89 |
ENSDART00000167842
|
si:dkey-72l17.5
|
si:dkey-72l17.5 |
| chr1_-_47071979 | 0.89 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr17_-_35076730 | 0.88 |
ENSDART00000146590
|
mboat2a
|
membrane bound O-acyltransferase domain containing 2a |
| chr16_+_29555801 | 0.88 |
ENSDART00000169425
|
ensab
|
endosulfine alpha b |
| chr19_+_1414604 | 0.87 |
ENSDART00000159024
|
btr29
|
bloodthirsty-related gene family, member 29 |
| chr16_-_25829779 | 0.87 |
ENSDART00000086301
|
irge4
|
immunity-related GTPase family, e4 |
| chr23_+_26142613 | 0.87 |
ENSDART00000165046
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr12_+_20641102 | 0.86 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr15_-_1003553 | 0.86 |
ENSDART00000154195
|
si:dkey-77f5.6
|
si:dkey-77f5.6 |
| chr13_+_7578111 | 0.85 |
ENSDART00000175431
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr15_-_29162193 | 0.85 |
ENSDART00000138449
ENSDART00000099885 |
xaf1
|
XIAP associated factor 1 |
| chr1_-_51266394 | 0.85 |
ENSDART00000164016
|
kif16ba
|
kinesin family member 16Ba |
| chr13_-_37620091 | 0.85 |
ENSDART00000135875
ENSDART00000193270 ENSDART00000018064 |
zgc:152791
|
zgc:152791 |
| chr12_-_4598237 | 0.83 |
ENSDART00000152489
|
irf3
|
interferon regulatory factor 3 |
| chr5_-_57723929 | 0.83 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr2_+_21090317 | 0.83 |
ENSDART00000109568
ENSDART00000139633 |
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr1_+_58424507 | 0.83 |
ENSDART00000114197
|
si:ch73-236c18.3
|
si:ch73-236c18.3 |
| chr4_+_34417403 | 0.83 |
ENSDART00000186032
ENSDART00000167909 |
si:ch211-246b8.2
|
si:ch211-246b8.2 |
| chr10_-_45229877 | 0.83 |
ENSDART00000169281
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr4_-_26095755 | 0.82 |
ENSDART00000100611
ENSDART00000191266 |
si:ch211-244b2.3
|
si:ch211-244b2.3 |
| chr15_+_17100697 | 0.82 |
ENSDART00000183565
ENSDART00000123197 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr7_-_72067475 | 0.82 |
ENSDART00000017763
|
LO018380.1
|
|
| chr13_+_17694845 | 0.81 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr5_+_8256574 | 0.81 |
ENSDART00000171343
|
FP102192.1
|
|
| chr15_+_2421729 | 0.80 |
ENSDART00000082294
ENSDART00000156428 |
hephl1a
|
hephaestin-like 1a |
| chr2_-_59376735 | 0.80 |
ENSDART00000193624
|
ftr38
|
finTRIM family, member 38 |
| chr1_-_12397258 | 0.80 |
ENSDART00000144596
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr15_-_20180475 | 0.79 |
ENSDART00000048183
|
exoc3l2b
|
exocyst complex component 3-like 2b |
| chr3_+_19446997 | 0.78 |
ENSDART00000079352
|
zgc:123297
|
zgc:123297 |
| chr21_+_34992550 | 0.78 |
ENSDART00000109041
ENSDART00000135400 |
tmprss15
|
transmembrane protease, serine 15 |
| chr5_+_52625975 | 0.78 |
ENSDART00000170341
ENSDART00000168317 |
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr2_-_59157790 | 0.77 |
ENSDART00000192303
ENSDART00000159362 |
ftr32
|
finTRIM family, member 32 |
| chr2_-_32688905 | 0.76 |
ENSDART00000041146
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr11_+_42478184 | 0.76 |
ENSDART00000089963
|
zgc:110286
|
zgc:110286 |
| chr15_-_47929455 | 0.76 |
ENSDART00000064462
|
psma6l
|
proteasome subunit alpha 6, like |
| chr2_-_30734098 | 0.76 |
ENSDART00000133769
|
rp1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr1_-_25177086 | 0.75 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr23_-_37835794 | 0.75 |
ENSDART00000137358
|
cd40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr23_-_12735434 | 0.75 |
ENSDART00000139166
|
si:dkey-96f10.1
|
si:dkey-96f10.1 |
| chr16_+_41570653 | 0.75 |
ENSDART00000102665
|
aste1a
|
asteroid homolog 1a |
| chr4_-_12795030 | 0.75 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr22_+_1615691 | 0.75 |
ENSDART00000171555
|
si:ch211-255f4.13
|
si:ch211-255f4.13 |
| chr12_-_26153101 | 0.74 |
ENSDART00000076051
|
opn4b
|
opsin 4b |
| chr11_+_2596667 | 0.74 |
ENSDART00000175330
|
dnajc14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr1_-_2457546 | 0.74 |
ENSDART00000103795
|
ggact.1
|
gamma-glutamylamine cyclotransferase, tandem duplicate 1 |
| chr12_+_27285994 | 0.73 |
ENSDART00000087204
|
dusp3a
|
dual specificity phosphatase 3a |
| chr8_-_38616712 | 0.73 |
ENSDART00000141827
|
si:ch211-198d23.1
|
si:ch211-198d23.1 |
| chr17_-_23631400 | 0.73 |
ENSDART00000079563
|
fas
|
Fas cell surface death receptor |
| chr9_-_19699728 | 0.73 |
ENSDART00000166780
|
si:ch211-141e20.2
|
si:ch211-141e20.2 |
| chr8_-_36469117 | 0.73 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr22_-_6054749 | 0.72 |
ENSDART00000191603
|
CU468041.2
|
|
| chr18_-_38270077 | 0.71 |
ENSDART00000185546
|
caprin1b
|
cell cycle associated protein 1b |
| chr1_+_7956030 | 0.70 |
ENSDART00000159655
|
CR855320.1
|
|
| chr18_+_13248956 | 0.70 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr10_-_7913591 | 0.69 |
ENSDART00000139661
|
slc35e4
|
solute carrier family 35, member E4 |
| chr22_-_5958066 | 0.69 |
ENSDART00000145821
|
si:rp71-36a1.3
|
si:rp71-36a1.3 |
| chr2_+_42005475 | 0.69 |
ENSDART00000056461
|
gbp2
|
guanylate binding protein 2 |
| chr7_+_8324506 | 0.69 |
ENSDART00000168110
|
si:dkey-185m8.2
|
si:dkey-185m8.2 |
| chr1_-_36771712 | 0.69 |
ENSDART00000148386
|
prmt9
|
protein arginine methyltransferase 9 |
| chr16_+_53710416 | 0.69 |
ENSDART00000155929
|
nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr22_-_29242347 | 0.68 |
ENSDART00000040761
|
pvalb7
|
parvalbumin 7 |
| chr7_-_6444011 | 0.68 |
ENSDART00000173010
|
zgc:112234
|
zgc:112234 |
| chr22_+_31023205 | 0.68 |
ENSDART00000111561
|
zmp:0000000735
|
zmp:0000000735 |
| chr23_-_12906228 | 0.68 |
ENSDART00000138807
|
ndnl2
|
necdin-like 2 |
| chr17_+_35362851 | 0.67 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr4_-_76079075 | 0.67 |
ENSDART00000158469
|
si:ch211-232d10.3
|
si:ch211-232d10.3 |
| chr16_+_26439939 | 0.67 |
ENSDART00000143073
|
trim35-28
|
tripartite motif containing 35-28 |
| chr1_+_40148193 | 0.67 |
ENSDART00000138072
|
zgc:194443
|
zgc:194443 |
| chr12_-_11560794 | 0.66 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr17_-_35361322 | 0.66 |
ENSDART00000019617
|
rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr20_+_4793516 | 0.66 |
ENSDART00000053875
|
lgals8a
|
galectin 8a |
| chr16_+_48678655 | 0.66 |
ENSDART00000150156
|
tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr5_-_38179220 | 0.66 |
ENSDART00000147701
|
si:ch211-284e13.11
|
si:ch211-284e13.11 |
| chr13_+_7387822 | 0.66 |
ENSDART00000148240
|
exoc3l4
|
exocyst complex component 3-like 4 |
| chr5_+_38662125 | 0.65 |
ENSDART00000136949
|
si:dkey-58f10.13
|
si:dkey-58f10.13 |
| chr2_+_9757453 | 0.64 |
ENSDART00000168972
|
pcyt1aa
|
phosphate cytidylyltransferase 1, choline, alpha a |
| chr15_-_37867995 | 0.64 |
ENSDART00000192698
|
si:dkey-238d18.4
|
si:dkey-238d18.4 |
| chr4_-_26108053 | 0.64 |
ENSDART00000066951
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr23_+_46183410 | 0.64 |
ENSDART00000167596
ENSDART00000151149 ENSDART00000150896 |
btr31
|
bloodthirsty-related gene family, member 31 |
| chr14_-_45967981 | 0.64 |
ENSDART00000188062
|
macrod1
|
MACRO domain containing 1 |
| chr4_+_53268458 | 0.64 |
ENSDART00000165335
|
si:dkey-250k10.1
|
si:dkey-250k10.1 |
| chr3_-_24093144 | 0.63 |
ENSDART00000103982
|
nfe2l1a
|
nuclear factor, erythroid 2-like 1a |
| chr3_-_4455951 | 0.63 |
ENSDART00000193908
ENSDART00000074077 |
trim35-3
|
tripartite motif containing 35-3 |
| chr23_+_21566828 | 0.63 |
ENSDART00000134741
ENSDART00000111966 |
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr20_+_715739 | 0.63 |
ENSDART00000136768
|
myo6a
|
myosin VIa |
| chr2_+_26656283 | 0.63 |
ENSDART00000133202
ENSDART00000099208 |
asph
|
aspartate beta-hydroxylase |
| chr15_-_36369743 | 0.63 |
ENSDART00000155942
|
si:dkey-23k10.3
|
si:dkey-23k10.3 |
| chr5_-_4923473 | 0.62 |
ENSDART00000134585
|
zbtb43
|
zinc finger and BTB domain containing 43 |
| chr16_-_51271962 | 0.62 |
ENSDART00000164021
ENSDART00000046420 |
serpinb1l1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 1 |
| chr25_+_245018 | 0.62 |
ENSDART00000155344
|
zgc:92481
|
zgc:92481 |
| chr15_-_36357889 | 0.61 |
ENSDART00000156377
|
si:dkey-23k10.5
|
si:dkey-23k10.5 |
| chr18_-_38270596 | 0.61 |
ENSDART00000098889
|
caprin1b
|
cell cycle associated protein 1b |
| chr9_+_12948511 | 0.60 |
ENSDART00000135797
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr2_-_47870649 | 0.59 |
ENSDART00000142854
|
ftr05
|
finTRIM family, member 5 |
| chr5_+_38619813 | 0.59 |
ENSDART00000133314
|
si:ch211-271e10.3
|
si:ch211-271e10.3 |
| chr19_+_23296616 | 0.58 |
ENSDART00000134567
|
irgf1
|
immunity-related GTPase family, f1 |
| chr3_-_43821381 | 0.58 |
ENSDART00000166021
|
snn
|
stannin |
| chr4_-_17669881 | 0.58 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr5_-_21030934 | 0.58 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr4_+_68476490 | 0.57 |
ENSDART00000168543
|
si:dkey-237g15.2
|
si:dkey-237g15.2 |
| chr22_-_6941098 | 0.57 |
ENSDART00000105864
|
zgc:171500
|
zgc:171500 |
| chr17_+_50524573 | 0.57 |
ENSDART00000187974
|
CR382385.1
|
|
| chr12_+_31608905 | 0.57 |
ENSDART00000152874
ENSDART00000152996 |
CR352342.1
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr3_-_336299 | 0.57 |
ENSDART00000105021
|
mhc1zfa
|
major histocompatibility complex class I ZFA |
| chr9_+_51147764 | 0.57 |
ENSDART00000188480
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr4_+_70967639 | 0.56 |
ENSDART00000159230
|
si:dkeyp-80d11.12
|
si:dkeyp-80d11.12 |
| chr8_-_51404806 | 0.56 |
ENSDART00000060625
|
lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr13_-_42066299 | 0.56 |
ENSDART00000111536
|
rmdn2
|
regulator of microtubule dynamics 2 |
| chr4_-_50933168 | 0.56 |
ENSDART00000150296
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr8_+_1187928 | 0.56 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr1_+_11881559 | 0.55 |
ENSDART00000166981
|
snx8b
|
sorting nexin 8b |
| chr3_-_40136743 | 0.55 |
ENSDART00000149546
|
myo15aa
|
myosin XVAa |
| chr3_-_32472155 | 0.55 |
ENSDART00000156280
|
si:ch211-195b15.7
|
si:ch211-195b15.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irf1b+irf2+irf2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 1.6 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.4 | 1.6 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.3 | 1.4 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.2 | 0.7 | GO:0045191 | response to protozoan(GO:0001562) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) defense response to protozoan(GO:0042832) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.2 | 1.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 2.0 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 0.9 | GO:0032728 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 0.7 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.2 | 1.9 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.2 | 1.4 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.2 | 0.9 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 0.9 | GO:0014005 | microglia development(GO:0014005) |
| 0.2 | 0.7 | GO:0070227 | lymphocyte apoptotic process(GO:0070227) |
| 0.2 | 0.9 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 1.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.2 | 0.5 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.2 | 1.1 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.2 | 0.7 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.2 | 1.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 1.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.8 | GO:0044819 | mitotic G1/S transition checkpoint(GO:0044819) |
| 0.1 | 1.0 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.4 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 0.6 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.9 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 0.4 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 1.6 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 0.3 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 0.3 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.4 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 1.4 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 0.3 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 0.3 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.8 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.2 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 1.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.7 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.4 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.1 | 1.5 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.9 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.3 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 | 0.9 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.4 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.7 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 | 2.2 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.1 | 0.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.4 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 1.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.4 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.1 | 0.6 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.1 | 0.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.3 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.1 | 0.3 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 0.5 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.5 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.0 | 0.3 | GO:0051793 | medium-chain fatty acid metabolic process(GO:0051791) medium-chain fatty acid catabolic process(GO:0051793) |
| 0.0 | 0.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 2.9 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.4 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.3 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.4 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 1.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 1.0 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 2.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 3.6 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.9 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.7 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.2 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.5 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.1 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 1.0 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.8 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.5 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.2 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.0 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 1.0 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 1.0 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.0 | 0.4 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.9 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.1 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.4 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.7 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0048660 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.1 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.5 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.1 | GO:0072088 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.0 | 1.2 | GO:0046474 | glycerophospholipid biosynthetic process(GO:0046474) |
| 0.0 | 0.0 | GO:0055057 | neuroblast division(GO:0055057) |
| 0.0 | 1.8 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.8 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.9 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 1.1 | GO:0046834 | lipid phosphorylation(GO:0046834) phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.2 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 1.0 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.3 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.5 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.7 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.4 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 0.8 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 1.1 | GO:0036503 | ERAD pathway(GO:0036503) |
| 0.0 | 1.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.4 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.3 | 1.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 1.5 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.2 | 1.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 1.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 0.7 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 1.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 0.8 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 1.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 3.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.4 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.9 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.4 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.3 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.3 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.9 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.9 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 4.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.3 | 2.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 0.9 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.3 | 1.9 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.2 | 0.7 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 1.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.2 | 0.8 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.2 | 1.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.2 | 1.6 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 0.8 | GO:0046977 | peptide antigen-transporting ATPase activity(GO:0015433) peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) |
| 0.2 | 0.6 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.6 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.1 | 2.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.6 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.4 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.7 | GO:1900750 | oligopeptide binding(GO:1900750) |
| 0.1 | 0.9 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.4 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.4 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.7 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 2.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.2 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 2.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.9 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 1.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.1 | GO:0047760 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.4 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.0 | 1.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 1.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.9 | GO:0016917 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 2.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.8 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 1.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 4.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.8 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 1.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.0 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 4.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 2.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 2.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.9 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 0.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 2.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |