Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for ikzf2
Z-value: 0.69
Transcription factors associated with ikzf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
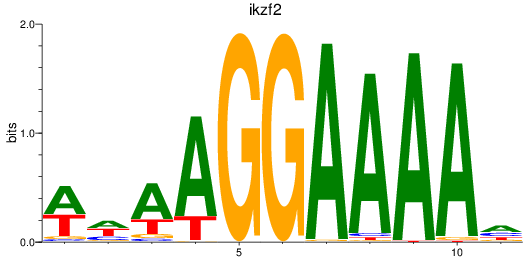
ikzf2
|
ENSDARG00000069111 | IKAROS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | dr11_v1_chr9_-_40073255_40073255 | -0.54 | 1.7e-02 | Click! |
Activity profile of ikzf2 motif
Sorted Z-values of ikzf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_26304386 | 1.45 |
ENSDART00000175416
|
otos
|
otospiralin |
| chr7_+_39386982 | 1.44 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr3_-_61203203 | 1.30 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr17_+_20204122 | 1.30 |
ENSDART00000078672
|
gnrh3
|
gonadotropin-releasing hormone 3 |
| chr2_+_45191049 | 1.29 |
ENSDART00000165392
|
ccl20a.3
|
chemokine (C-C motif) ligand 20a, duplicate 3 |
| chr12_-_30844304 | 1.28 |
ENSDART00000146367
|
crygmxl2
|
crystallin, gamma MX, like 2 |
| chr21_+_25187210 | 1.28 |
ENSDART00000101147
ENSDART00000167528 |
si:dkey-183i3.5
|
si:dkey-183i3.5 |
| chr25_-_18470695 | 1.26 |
ENSDART00000034377
|
cpa5
|
carboxypeptidase A5 |
| chr22_-_20011476 | 1.23 |
ENSDART00000093312
ENSDART00000093310 |
celf5a
|
cugbp, Elav-like family member 5a |
| chr23_-_32162810 | 1.17 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr11_+_25459697 | 1.12 |
ENSDART00000161481
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr2_-_37210397 | 1.12 |
ENSDART00000084938
|
apoda.1
|
apolipoprotein Da, duplicate 1 |
| chr20_-_20821783 | 1.03 |
ENSDART00000152577
ENSDART00000027603 ENSDART00000145601 |
ckbb
|
creatine kinase, brain b |
| chr25_+_15841670 | 0.96 |
ENSDART00000049992
|
syt9b
|
synaptotagmin IXb |
| chr15_-_27710513 | 0.93 |
ENSDART00000005641
ENSDART00000134373 |
lhx1a
|
LIM homeobox 1a |
| chr13_+_22264914 | 0.90 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr5_-_33215261 | 0.90 |
ENSDART00000097935
ENSDART00000134777 |
si:dkey-226m8.10
|
si:dkey-226m8.10 |
| chr6_-_48347498 | 0.89 |
ENSDART00000149159
|
capza1a
|
capping protein (actin filament) muscle Z-line, alpha 1a |
| chr12_-_15205087 | 0.87 |
ENSDART00000010068
|
sult1st6
|
sulfotransferase family 1, cytosolic sulfotransferase 6 |
| chr25_-_8602437 | 0.86 |
ENSDART00000171200
|
rhcgb
|
Rh family, C glycoprotein b |
| chr1_-_56223913 | 0.86 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr6_-_47246948 | 0.85 |
ENSDART00000162435
|
grm4
|
glutamate receptor, metabotropic 4 |
| chr22_+_20427170 | 0.83 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr3_+_32142382 | 0.82 |
ENSDART00000133035
|
syt5a
|
synaptotagmin Va |
| chr3_-_61181018 | 0.80 |
ENSDART00000187970
|
pvalb4
|
parvalbumin 4 |
| chr16_-_12914288 | 0.79 |
ENSDART00000184221
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr12_-_4841018 | 0.79 |
ENSDART00000166500
|
zgc:163073
|
zgc:163073 |
| chr6_+_38381957 | 0.79 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr2_-_37465517 | 0.77 |
ENSDART00000139983
|
si:dkey-57k2.6
|
si:dkey-57k2.6 |
| chr18_+_8346920 | 0.77 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr5_-_65158203 | 0.76 |
ENSDART00000171656
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr18_-_21177674 | 0.74 |
ENSDART00000060175
|
si:dkey-12e7.4
|
si:dkey-12e7.4 |
| chr22_-_16412496 | 0.70 |
ENSDART00000137497
|
cts12
|
cathepsin 12 |
| chr18_-_17485419 | 0.69 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr11_-_32723851 | 0.67 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr5_+_36932718 | 0.66 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr3_+_39540014 | 0.65 |
ENSDART00000074848
|
zgc:165423
|
zgc:165423 |
| chr9_+_50007556 | 0.64 |
ENSDART00000175587
|
slc38a11
|
solute carrier family 38, member 11 |
| chr7_+_13382852 | 0.64 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr20_+_46040666 | 0.63 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr13_-_349952 | 0.62 |
ENSDART00000133731
ENSDART00000140326 ENSDART00000189389 ENSDART00000185865 ENSDART00000109634 ENSDART00000147058 ENSDART00000142695 |
si:ch1073-291c23.2
|
si:ch1073-291c23.2 |
| chr17_-_15747296 | 0.62 |
ENSDART00000153970
|
cx52.7
|
connexin 52.7 |
| chr4_+_9279784 | 0.62 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr19_-_31522625 | 0.61 |
ENSDART00000158438
ENSDART00000035049 |
necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr5_-_38506981 | 0.60 |
ENSDART00000097822
|
atp1b2b
|
ATPase Na+/K+ transporting subunit beta 2b |
| chr2_-_13216269 | 0.60 |
ENSDART00000149947
|
bcl2b
|
BCL2, apoptosis regulator b |
| chr2_-_32643738 | 0.60 |
ENSDART00000112452
|
si:dkeyp-73d8.9
|
si:dkeyp-73d8.9 |
| chr18_+_39074139 | 0.58 |
ENSDART00000142390
|
gnb5b
|
guanine nucleotide binding protein (G protein), beta 5b |
| chr20_+_16881883 | 0.57 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr15_-_37829160 | 0.57 |
ENSDART00000099425
|
ctrl
|
chymotrypsin-like |
| chr1_-_26292897 | 0.56 |
ENSDART00000112899
ENSDART00000185410 |
cxxc4
|
CXXC finger 4 |
| chr14_-_3381303 | 0.56 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr4_+_7508316 | 0.56 |
ENSDART00000170924
ENSDART00000170933 ENSDART00000164985 ENSDART00000167571 ENSDART00000158843 ENSDART00000158999 |
tnnt2e
|
troponin T2e, cardiac |
| chr8_+_1148876 | 0.56 |
ENSDART00000148651
|
avp
|
arginine vasopressin |
| chr23_-_20764227 | 0.54 |
ENSDART00000089750
|
znf362b
|
zinc finger protein 362b |
| chr25_-_8160030 | 0.54 |
ENSDART00000067159
|
tph1a
|
tryptophan hydroxylase 1 (tryptophan 5-monooxygenase) a |
| chr12_+_28367557 | 0.53 |
ENSDART00000066294
|
cdk5r1b
|
cyclin-dependent kinase 5, regulatory subunit 1b (p35) |
| chr23_+_20408227 | 0.53 |
ENSDART00000134727
|
si:rp71-17i16.4
|
si:rp71-17i16.4 |
| chr5_-_68916623 | 0.53 |
ENSDART00000141917
ENSDART00000109053 |
ank1a
|
ankyrin 1, erythrocytic a |
| chr21_+_27189490 | 0.52 |
ENSDART00000125349
|
bada
|
BCL2 associated agonist of cell death a |
| chr2_+_55984788 | 0.52 |
ENSDART00000183599
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr20_-_5291012 | 0.52 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr19_-_40985308 | 0.51 |
ENSDART00000132986
ENSDART00000142360 |
si:ch211-120e1.7
|
si:ch211-120e1.7 |
| chr11_-_13126505 | 0.51 |
ENSDART00000158377
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr14_-_33348221 | 0.50 |
ENSDART00000187749
|
rpl39
|
ribosomal protein L39 |
| chr1_+_15137901 | 0.49 |
ENSDART00000111475
|
pcdh7a
|
protocadherin 7a |
| chr17_-_28749640 | 0.49 |
ENSDART00000000948
|
coch
|
coagulation factor C homolog, cochlin (Limulus polyphemus) |
| chr20_+_34717403 | 0.49 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr7_+_46003449 | 0.49 |
ENSDART00000159700
ENSDART00000173625 |
si:ch211-260e23.9
|
si:ch211-260e23.9 |
| chr15_-_19250543 | 0.48 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr18_-_16123222 | 0.48 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr23_-_24450686 | 0.48 |
ENSDART00000189161
|
spen
|
spen family transcriptional repressor |
| chr2_-_22535 | 0.48 |
ENSDART00000157877
|
CABZ01092282.1
|
|
| chr16_-_33095161 | 0.47 |
ENSDART00000187648
|
dopey1
|
dopey family member 1 |
| chr2_-_31735142 | 0.47 |
ENSDART00000130903
|
ralyl
|
RALY RNA binding protein like |
| chr12_+_17106117 | 0.47 |
ENSDART00000149990
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr1_+_2712956 | 0.46 |
ENSDART00000126093
|
gpc6a
|
glypican 6a |
| chr23_+_44883805 | 0.45 |
ENSDART00000182805
|
si:ch73-361h17.1
|
si:ch73-361h17.1 |
| chr8_+_20880848 | 0.45 |
ENSDART00000134488
ENSDART00000138605 ENSDART00000192234 |
si:ch73-196i15.3
|
si:ch73-196i15.3 |
| chr3_-_28428198 | 0.45 |
ENSDART00000151546
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr9_-_22057658 | 0.44 |
ENSDART00000101944
|
crygmxl1
|
crystallin, gamma MX, like 1 |
| chr8_-_52413032 | 0.44 |
ENSDART00000183039
|
CABZ01070469.1
|
|
| chr12_-_48477031 | 0.44 |
ENSDART00000105176
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr7_-_39551913 | 0.44 |
ENSDART00000173498
ENSDART00000143040 |
slc22a18
|
solute carrier family 22, member 18 |
| chr5_-_68916455 | 0.43 |
ENSDART00000171465
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr12_-_19007834 | 0.43 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr4_+_21129752 | 0.42 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr24_+_34085940 | 0.42 |
ENSDART00000171189
|
asb10
|
ankyrin repeat and SOCS box containing 10 |
| chr3_+_55131289 | 0.42 |
ENSDART00000111585
|
AL935210.1
|
|
| chr18_+_783936 | 0.42 |
ENSDART00000193357
|
rpp25b
|
ribonuclease P and MRP subunit p25, b |
| chr3_-_44059902 | 0.42 |
ENSDART00000158485
ENSDART00000159088 ENSDART00000165628 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr16_-_9980402 | 0.42 |
ENSDART00000066372
|
id4
|
inhibitor of DNA binding 4 |
| chr19_+_29798064 | 0.41 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr5_+_20428838 | 0.41 |
ENSDART00000141118
|
tmem119a
|
transmembrane protein 119a |
| chr18_+_17663898 | 0.41 |
ENSDART00000021213
|
cpne2
|
copine II |
| chr14_-_21219659 | 0.40 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr25_+_33939728 | 0.40 |
ENSDART00000148537
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr9_+_14010823 | 0.40 |
ENSDART00000143837
|
si:ch211-67e16.3
|
si:ch211-67e16.3 |
| chr21_-_37889727 | 0.40 |
ENSDART00000163612
ENSDART00000180958 |
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr22_-_28650442 | 0.40 |
ENSDART00000019846
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr22_-_11137268 | 0.39 |
ENSDART00000178882
|
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr14_-_32876280 | 0.39 |
ENSDART00000173168
|
si:rp71-46j2.7
|
si:rp71-46j2.7 |
| chr20_+_7799615 | 0.39 |
ENSDART00000152792
|
si:dkey-19f4.2
|
si:dkey-19f4.2 |
| chr7_-_16562200 | 0.39 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr21_+_11468642 | 0.39 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr24_-_12689571 | 0.39 |
ENSDART00000015517
|
pdcd6
|
programmed cell death 6 |
| chr4_-_72609735 | 0.38 |
ENSDART00000174299
ENSDART00000159227 |
si:cabz01054394.6
|
si:cabz01054394.6 |
| chr8_-_14049404 | 0.38 |
ENSDART00000093117
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr25_-_35960229 | 0.38 |
ENSDART00000073434
|
snx20
|
sorting nexin 20 |
| chr12_+_24344611 | 0.38 |
ENSDART00000093094
|
nrxn1a
|
neurexin 1a |
| chr13_+_36680564 | 0.38 |
ENSDART00000136030
ENSDART00000006923 |
atp5s
|
ATP synthase, H+ transporting, mitochondrial F0 complex subunit s |
| chr24_+_29449690 | 0.37 |
ENSDART00000105743
ENSDART00000193556 ENSDART00000145816 |
ntng1a
|
netrin g1a |
| chr13_+_16521898 | 0.36 |
ENSDART00000122557
|
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr5_-_17876709 | 0.36 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr21_-_33851877 | 0.36 |
ENSDART00000136729
|
ebf1b
|
early B cell factor 1b |
| chr15_-_46779934 | 0.36 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr18_+_7591381 | 0.36 |
ENSDART00000136313
|
si:dkeyp-1h4.6
|
si:dkeyp-1h4.6 |
| chr6_-_31348999 | 0.35 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr6_+_39360377 | 0.35 |
ENSDART00000028260
ENSDART00000151322 |
zgc:77517
|
zgc:77517 |
| chr22_+_15310103 | 0.35 |
ENSDART00000145849
|
CU104797.1
|
|
| chr10_+_41765944 | 0.35 |
ENSDART00000171484
|
rnf34b
|
ring finger protein 34b |
| chr4_+_76659013 | 0.35 |
ENSDART00000147908
ENSDART00000134229 |
ms4a17a.5
|
membrane-spanning 4-domains, subfamily A, member 17A.5 |
| chr22_+_5106751 | 0.35 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr7_-_31940590 | 0.35 |
ENSDART00000131009
|
bdnf
|
brain-derived neurotrophic factor |
| chr16_+_19732543 | 0.35 |
ENSDART00000149901
ENSDART00000052927 |
twist1b
|
twist family bHLH transcription factor 1b |
| chr7_-_35432901 | 0.35 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr6_-_13475439 | 0.34 |
ENSDART00000172318
|
mylkb
|
myosin light chain kinase b |
| chr7_-_23971497 | 0.34 |
ENSDART00000173603
|
si:dkey-183c6.8
|
si:dkey-183c6.8 |
| chr12_-_9516981 | 0.34 |
ENSDART00000106285
|
si:ch211-207i20.3
|
si:ch211-207i20.3 |
| chr24_-_11905911 | 0.34 |
ENSDART00000033621
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr5_+_23624684 | 0.33 |
ENSDART00000051539
|
cx27.5
|
connexin 27.5 |
| chr12_+_14084291 | 0.33 |
ENSDART00000189734
|
si:ch211-217a12.1
|
si:ch211-217a12.1 |
| chr17_+_19626479 | 0.33 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr21_+_37445871 | 0.32 |
ENSDART00000076333
|
pgk1
|
phosphoglycerate kinase 1 |
| chr5_+_4332220 | 0.32 |
ENSDART00000051699
|
sat1a.1
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 1 |
| chr1_+_9966384 | 0.32 |
ENSDART00000132607
|
si:dkeyp-75b4.8
|
si:dkeyp-75b4.8 |
| chr9_+_45839260 | 0.31 |
ENSDART00000114814
|
twist2
|
twist2 |
| chr7_+_73822054 | 0.31 |
ENSDART00000109720
|
zgc:163061
|
zgc:163061 |
| chr9_+_32069989 | 0.31 |
ENSDART00000139540
|
si:dkey-83m22.7
|
si:dkey-83m22.7 |
| chr21_-_21555039 | 0.31 |
ENSDART00000143031
|
or133-5
|
odorant receptor, family H, subfamily 133, member 5 |
| chr5_-_13835461 | 0.31 |
ENSDART00000148297
ENSDART00000114841 |
add2
|
adducin 2 (beta) |
| chr7_+_69470442 | 0.31 |
ENSDART00000189593
|
gabarapb
|
GABA(A) receptor-associated protein b |
| chr10_+_36662640 | 0.31 |
ENSDART00000063359
|
ucp2
|
uncoupling protein 2 |
| chr25_+_5039050 | 0.31 |
ENSDART00000154700
|
parvb
|
parvin, beta |
| chr15_+_15314189 | 0.30 |
ENSDART00000156830
|
camkk1b
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha b |
| chr11_+_34235372 | 0.30 |
ENSDART00000063150
|
fam43a
|
family with sequence similarity 43, member A |
| chr14_+_33413980 | 0.30 |
ENSDART00000052780
ENSDART00000124437 ENSDART00000173327 |
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr17_+_8211992 | 0.30 |
ENSDART00000133301
|
slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr21_+_11468934 | 0.30 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr12_+_24344963 | 0.29 |
ENSDART00000191648
ENSDART00000183180 ENSDART00000088178 ENSDART00000189696 |
nrxn1a
|
neurexin 1a |
| chr14_-_4043818 | 0.29 |
ENSDART00000179870
|
snx25
|
sorting nexin 25 |
| chr2_-_48196092 | 0.29 |
ENSDART00000139944
|
snorc
|
secondary ossification center associated regulator of chondrocyte maturation |
| chr24_-_39858710 | 0.29 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr14_+_30774515 | 0.29 |
ENSDART00000191666
|
atl3
|
atlastin 3 |
| chr11_-_2469529 | 0.29 |
ENSDART00000125677
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr13_+_16522608 | 0.28 |
ENSDART00000182838
ENSDART00000143200 |
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr19_-_32804535 | 0.28 |
ENSDART00000175613
ENSDART00000052098 |
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr21_+_11248448 | 0.28 |
ENSDART00000142431
|
arid6
|
AT-rich interaction domain 6 |
| chr16_-_31976269 | 0.28 |
ENSDART00000139664
|
styk1
|
serine/threonine/tyrosine kinase 1 |
| chr17_-_15746898 | 0.28 |
ENSDART00000180079
|
cx52.7
|
connexin 52.7 |
| chr10_+_21758811 | 0.28 |
ENSDART00000188827
|
pcdh1g11
|
protocadherin 1 gamma 11 |
| chr2_-_36925561 | 0.28 |
ENSDART00000187690
|
map1sb
|
microtubule-associated protein 1Sb |
| chr6_-_46742455 | 0.27 |
ENSDART00000011970
|
zgc:66479
|
zgc:66479 |
| chr19_-_26869103 | 0.27 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr10_-_36808348 | 0.27 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr4_+_18843015 | 0.27 |
ENSDART00000152086
ENSDART00000066977 ENSDART00000132567 |
bik
|
BCL2 interacting killer |
| chr7_+_29009161 | 0.27 |
ENSDART00000166458
|
CR376738.1
|
|
| chr15_+_8043751 | 0.27 |
ENSDART00000193701
|
cadm2b
|
cell adhesion molecule 2b |
| chr16_-_38333976 | 0.27 |
ENSDART00000031895
|
cdc42se1
|
CDC42 small effector 1 |
| chr1_-_1631399 | 0.27 |
ENSDART00000176787
|
clic6
|
chloride intracellular channel 6 |
| chr6_-_8489810 | 0.26 |
ENSDART00000124643
|
rasal3
|
RAS protein activator like 3 |
| chr17_+_33340675 | 0.26 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr11_+_10548171 | 0.26 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr12_-_32066469 | 0.26 |
ENSDART00000140685
ENSDART00000062185 |
rab40b
|
RAB40B, member RAS oncogene family |
| chr21_+_40232910 | 0.26 |
ENSDART00000075915
|
or115-11
|
odorant receptor, family F, subfamily 115, member 11 |
| chr19_+_46222918 | 0.26 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr23_+_19675780 | 0.26 |
ENSDART00000124335
|
smim4
|
small integral membrane protein 4 |
| chr25_-_13188214 | 0.25 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr13_-_30700460 | 0.25 |
ENSDART00000139073
|
rassf4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr12_+_42436920 | 0.25 |
ENSDART00000177303
|
ebf3a
|
early B cell factor 3a |
| chr14_+_30774032 | 0.25 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr11_+_21910343 | 0.25 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr14_-_14566417 | 0.24 |
ENSDART00000159056
|
si:dkey-27i16.2
|
si:dkey-27i16.2 |
| chr5_+_32882688 | 0.24 |
ENSDART00000008807
ENSDART00000185317 |
rpl12
|
ribosomal protein L12 |
| chr19_+_46222428 | 0.23 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr6_+_29791164 | 0.23 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr9_+_11202552 | 0.23 |
ENSDART00000151931
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr10_-_5857548 | 0.23 |
ENSDART00000166933
|
si:ch211-281k19.2
|
si:ch211-281k19.2 |
| chr12_+_22576404 | 0.23 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr10_+_21737745 | 0.23 |
ENSDART00000170498
ENSDART00000167997 |
pcdh1g18
|
protocadherin 1 gamma 18 |
| chr6_+_8176486 | 0.23 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr7_+_2228276 | 0.23 |
ENSDART00000064294
|
si:dkey-187j14.4
|
si:dkey-187j14.4 |
| chr2_-_31634978 | 0.23 |
ENSDART00000135668
|
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr13_-_31397987 | 0.23 |
ENSDART00000008287
|
pgam1a
|
phosphoglycerate mutase 1a |
| chr11_-_17964525 | 0.22 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr24_-_18179535 | 0.22 |
ENSDART00000186112
|
cntnap2a
|
contactin associated protein like 2a |
| chr12_+_41991635 | 0.22 |
ENSDART00000186161
ENSDART00000192510 |
TCERG1L
|
transcription elongation regulator 1 like |
| chr12_-_4070058 | 0.22 |
ENSDART00000042200
|
aldoab
|
aldolase a, fructose-bisphosphate, b |
| chr7_+_19482084 | 0.22 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr8_+_26859639 | 0.22 |
ENSDART00000133440
|
prdm2a
|
PR domain containing 2, with ZNF domain a |
Network of associatons between targets according to the STRING database.
First level regulatory network of ikzf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0090189 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.2 | 0.6 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.2 | 0.9 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.2 | 0.6 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.2 | 0.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.4 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.1 | 1.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.4 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.1 | 0.4 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 1.2 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.5 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.3 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.6 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.3 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.2 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.1 | 1.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 1.1 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 1.1 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.6 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.4 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.6 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.0 | 0.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.8 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.3 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.2 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.1 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.8 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.2 | GO:0051701 | interaction with host(GO:0051701) |
| 0.0 | 0.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.3 | GO:0009118 | regulation of oxidative phosphorylation(GO:0002082) regulation of nucleoside metabolic process(GO:0009118) regulation of ATP metabolic process(GO:1903578) |
| 0.0 | 0.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 2.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.2 | GO:2000583 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.0 | 0.2 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.1 | GO:0042942 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.0 | 0.3 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.2 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 1.1 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.4 | GO:0003307 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) |
| 0.0 | 1.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.2 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.0 | 0.1 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 1.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.9 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 1.4 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.3 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.1 | GO:0009886 | cardiac conduction system development(GO:0003161) post-embryonic morphogenesis(GO:0009886) |
| 0.0 | 0.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.6 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.2 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.4 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.5 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.7 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 1.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.3 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.6 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.9 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.3 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0030826 | nitric oxide mediated signal transduction(GO:0007263) regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 1.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.0 | 0.0 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.0 | 0.3 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.3 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.3 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.0 | 0.1 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 0.9 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.9 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.6 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 2.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.1 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.0 | GO:0033267 | axon part(GO:0033267) |
| 0.0 | 1.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.0 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.1 | 0.8 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.5 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 1.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.9 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.3 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.5 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.1 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.5 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.2 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.1 | 0.2 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.6 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 2.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.8 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.6 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.9 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 1.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 1.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:1990931 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.3 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 1.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 1.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.0 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.0 | GO:0070513 | death domain binding(GO:0070513) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.1 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |