Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
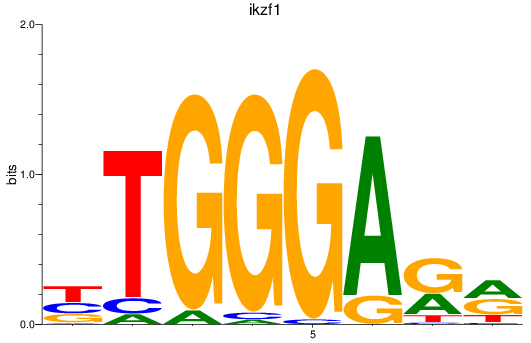
Results for ikzf1
Z-value: 0.94
Transcription factors associated with ikzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ikzf1
|
ENSDARG00000013539 | IKAROS family zinc finger 1 (Ikaros) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ikzf1 | dr11_v1_chr13_-_15986871_15986871 | -0.78 | 7.6e-05 | Click! |
Activity profile of ikzf1 motif
Sorted Z-values of ikzf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_29161609 | 3.30 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr7_-_8416750 | 2.05 |
ENSDART00000181857
|
jac1
|
jacalin 1 |
| chr5_-_41831646 | 1.93 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr17_+_25414033 | 1.90 |
ENSDART00000001691
|
tdh2
|
L-threonine dehydrogenase 2 |
| chr5_-_71722257 | 1.90 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr21_+_7582036 | 1.74 |
ENSDART00000135485
ENSDART00000027268 |
otpa
|
orthopedia homeobox a |
| chr7_+_39386982 | 1.63 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr3_+_25154078 | 1.61 |
ENSDART00000156973
|
si:ch211-256m1.8
|
si:ch211-256m1.8 |
| chr2_-_34555945 | 1.59 |
ENSDART00000056671
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr5_+_32222303 | 1.55 |
ENSDART00000051362
|
myhc4
|
myosin heavy chain 4 |
| chr5_-_64883082 | 1.50 |
ENSDART00000064983
ENSDART00000139066 |
krt1-c5
|
keratin, type 1, gene c5 |
| chr20_+_20637866 | 1.43 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr16_+_31511739 | 1.42 |
ENSDART00000049420
|
ndrg1b
|
N-myc downstream regulated 1b |
| chr6_-_39764995 | 1.39 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr2_+_42724404 | 1.34 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr1_+_37391141 | 1.34 |
ENSDART00000083593
ENSDART00000168647 |
sparcl1
|
SPARC-like 1 |
| chr7_-_45076131 | 1.31 |
ENSDART00000110590
|
zgc:194678
|
zgc:194678 |
| chr20_+_20638034 | 1.30 |
ENSDART00000189759
|
rtn1b
|
reticulon 1b |
| chr3_-_32817274 | 1.29 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr13_-_2189761 | 1.27 |
ENSDART00000166255
|
mlip
|
muscular LMNA-interacting protein |
| chr2_-_33645411 | 1.26 |
ENSDART00000114663
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr3_-_59981162 | 1.15 |
ENSDART00000128790
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr19_-_9829965 | 1.14 |
ENSDART00000136842
ENSDART00000142766 |
cacng8a
|
calcium channel, voltage-dependent, gamma subunit 8a |
| chr1_+_135903 | 1.14 |
ENSDART00000124837
|
f10
|
coagulation factor X |
| chr10_+_21786656 | 1.13 |
ENSDART00000185851
ENSDART00000167219 |
pcdh1g26
|
protocadherin 1 gamma 26 |
| chr16_-_54455573 | 1.12 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr5_-_26181863 | 1.12 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr5_-_57641257 | 1.09 |
ENSDART00000149282
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr21_-_131236 | 1.09 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr16_+_50100420 | 1.08 |
ENSDART00000128167
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr9_-_5046315 | 1.08 |
ENSDART00000179087
ENSDART00000109954 |
nr4a2a
|
nuclear receptor subfamily 4, group A, member 2a |
| chr3_+_17806213 | 1.08 |
ENSDART00000055890
|
znf385c
|
zinc finger protein 385C |
| chr2_-_51794472 | 1.07 |
ENSDART00000186652
|
BX908782.3
|
|
| chr3_-_59981476 | 1.06 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr1_-_44434707 | 1.06 |
ENSDART00000110148
|
cryba1l2
|
crystallin, beta A1, like 2 |
| chr20_+_1398564 | 1.05 |
ENSDART00000002242
|
leg1.2
|
liver-enriched gene 1, tandem duplicate 2 |
| chr3_+_40170216 | 1.03 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr1_+_36436936 | 1.02 |
ENSDART00000124112
|
pou4f2
|
POU class 4 homeobox 2 |
| chr14_-_39031108 | 1.01 |
ENSDART00000026194
|
glra4a
|
glycine receptor, alpha 4a |
| chr2_+_6885852 | 1.00 |
ENSDART00000016607
|
rgs5b
|
regulator of G protein signaling 5b |
| chr2_-_31936966 | 1.00 |
ENSDART00000169484
ENSDART00000192492 ENSDART00000027689 |
amph
|
amphiphysin |
| chr21_-_10773344 | 1.00 |
ENSDART00000063244
|
grp
|
gastrin-releasing peptide |
| chr23_+_6795531 | 0.99 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr23_+_4288964 | 0.99 |
ENSDART00000138408
|
l3mbtl1
|
l(3)mbt-like 1 (Drosophila) |
| chr23_+_3616224 | 0.99 |
ENSDART00000190917
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr14_-_2196267 | 0.99 |
ENSDART00000161674
ENSDART00000125674 |
pcdh2ab8
pcdh2ab9
|
protocadherin 2 alpha b 8 protocadherin 2 alpha b 9 |
| chr2_-_30784502 | 0.98 |
ENSDART00000056735
|
rgs20
|
regulator of G protein signaling 20 |
| chr6_+_48618512 | 0.98 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr18_-_21177674 | 0.96 |
ENSDART00000060175
|
si:dkey-12e7.4
|
si:dkey-12e7.4 |
| chr5_+_6892195 | 0.96 |
ENSDART00000048201
|
gb:cr929477
|
expressed sequence CR929477 |
| chr1_+_36437585 | 0.96 |
ENSDART00000189182
|
pou4f2
|
POU class 4 homeobox 2 |
| chr14_-_51855047 | 0.95 |
ENSDART00000088912
|
cplx1
|
complexin 1 |
| chr25_-_19224298 | 0.95 |
ENSDART00000149917
|
acanb
|
aggrecan b |
| chr15_-_47193564 | 0.95 |
ENSDART00000172453
|
LSAMP
|
limbic system-associated membrane protein |
| chr3_-_52614747 | 0.94 |
ENSDART00000154365
|
trim35-13
|
tripartite motif containing 35-13 |
| chr7_+_61764040 | 0.91 |
ENSDART00000056745
|
acox3
|
acyl-CoA oxidase 3, pristanoyl |
| chr21_+_39100289 | 0.90 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr13_-_42306348 | 0.90 |
ENSDART00000003706
|
kmo
|
kynurenine 3-monooxygenase |
| chr17_+_2549503 | 0.89 |
ENSDART00000156843
|
si:dkey-248g15.3
|
si:dkey-248g15.3 |
| chr8_+_37465380 | 0.89 |
ENSDART00000003954
|
mapkapk2b
|
mitogen-activated protein kinase-activated protein kinase 2b |
| chr14_+_25817246 | 0.87 |
ENSDART00000136733
|
glra1
|
glycine receptor, alpha 1 |
| chr20_-_45040916 | 0.86 |
ENSDART00000190001
|
klhl29
|
kelch-like family member 29 |
| chr13_+_38302665 | 0.85 |
ENSDART00000145777
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr16_+_1353894 | 0.85 |
ENSDART00000148426
|
celf3b
|
cugbp, Elav-like family member 3b |
| chr5_+_23118470 | 0.85 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr19_-_47452874 | 0.83 |
ENSDART00000025931
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr11_+_24251141 | 0.81 |
ENSDART00000182684
|
pnp4a
|
purine nucleoside phosphorylase 4a |
| chr20_+_50852356 | 0.80 |
ENSDART00000167517
ENSDART00000168396 |
gphnb
|
gephyrin b |
| chr15_-_12229874 | 0.80 |
ENSDART00000165159
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr12_+_42574148 | 0.80 |
ENSDART00000157855
|
ebf3a
|
early B cell factor 3a |
| chr23_-_18287618 | 0.79 |
ENSDART00000112735
|
fam19a1a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1a |
| chr18_-_46135927 | 0.78 |
ENSDART00000134537
ENSDART00000191481 |
SPTBN4
|
si:ch73-262h23.4 |
| chr7_-_38792543 | 0.78 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr11_-_1967438 | 0.77 |
ENSDART00000155844
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr3_+_14768364 | 0.77 |
ENSDART00000090235
ENSDART00000139001 |
nfixb
|
nuclear factor I/Xb |
| chr24_+_13316737 | 0.77 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
| chr1_-_43712120 | 0.76 |
ENSDART00000074600
|
SLC9B2
|
si:dkey-162b23.4 |
| chr22_-_29191152 | 0.76 |
ENSDART00000132702
|
pvalb7
|
parvalbumin 7 |
| chr15_-_24869826 | 0.75 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr13_+_27951688 | 0.75 |
ENSDART00000050303
|
b3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr10_+_375042 | 0.75 |
ENSDART00000171854
|
si:ch1073-303d10.1
|
si:ch1073-303d10.1 |
| chr2_-_30784198 | 0.74 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr11_-_21030070 | 0.74 |
ENSDART00000186322
|
fmoda
|
fibromodulin a |
| chr6_+_9175886 | 0.72 |
ENSDART00000165333
|
si:ch211-207l14.1
|
si:ch211-207l14.1 |
| chr8_+_52642869 | 0.72 |
ENSDART00000163617
ENSDART00000189997 |
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr22_-_16412496 | 0.71 |
ENSDART00000137497
|
cts12
|
cathepsin 12 |
| chr21_-_39546737 | 0.71 |
ENSDART00000006971
|
sept4a
|
septin 4a |
| chr1_+_47446032 | 0.71 |
ENSDART00000126904
ENSDART00000007262 |
gja8b
|
gap junction protein alpha 8 paralog b |
| chr7_-_20731078 | 0.69 |
ENSDART00000188267
|
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr1_-_12160981 | 0.69 |
ENSDART00000162146
|
b4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr1_-_5455498 | 0.69 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr16_+_37470717 | 0.68 |
ENSDART00000112003
ENSDART00000188431 ENSDART00000192837 |
adgrb1a
|
adhesion G protein-coupled receptor B1a |
| chr7_-_8438657 | 0.67 |
ENSDART00000173054
|
si:dkeyp-32g11.8
|
si:dkeyp-32g11.8 |
| chr21_-_12119711 | 0.67 |
ENSDART00000131538
|
celf4
|
CUGBP, Elav-like family member 4 |
| chr1_-_26702930 | 0.67 |
ENSDART00000109297
ENSDART00000152389 |
foxe1
|
forkhead box E1 |
| chr5_-_41838354 | 0.67 |
ENSDART00000146793
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr6_+_2195625 | 0.66 |
ENSDART00000155659
|
acvr1bb
|
activin A receptor type 1Bb |
| chr12_-_15620090 | 0.66 |
ENSDART00000038032
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr3_-_58189429 | 0.66 |
ENSDART00000156092
|
si:ch211-256e16.11
|
si:ch211-256e16.11 |
| chr15_-_23376541 | 0.65 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr12_-_47782623 | 0.65 |
ENSDART00000115742
|
selenou1b
|
selenoprotein U1b |
| chr20_+_15982482 | 0.65 |
ENSDART00000020999
|
angptl1a
|
angiopoietin-like 1a |
| chr22_-_18164835 | 0.64 |
ENSDART00000143189
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr6_-_53204808 | 0.64 |
ENSDART00000160955
|
slc38a3b
|
solute carrier family 38, member 3b |
| chr25_-_31396479 | 0.64 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr18_-_46763170 | 0.63 |
ENSDART00000171880
|
dner
|
delta/notch-like EGF repeat containing |
| chr11_-_37548711 | 0.62 |
ENSDART00000127184
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr7_+_31879649 | 0.62 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr3_-_2591942 | 0.62 |
ENSDART00000127971
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr5_-_43071058 | 0.60 |
ENSDART00000165546
|
si:dkey-245n4.2
|
si:dkey-245n4.2 |
| chr15_-_8517376 | 0.60 |
ENSDART00000186289
|
npas1
|
neuronal PAS domain protein 1 |
| chr23_-_11870962 | 0.60 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr14_+_11762991 | 0.60 |
ENSDART00000110004
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr9_-_44642108 | 0.60 |
ENSDART00000086202
|
pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr7_-_58098814 | 0.59 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr2_-_15324837 | 0.59 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr17_+_48174381 | 0.59 |
ENSDART00000191721
ENSDART00000193225 ENSDART00000184542 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr23_-_3674443 | 0.59 |
ENSDART00000134830
ENSDART00000057422 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr23_+_6077503 | 0.58 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr14_+_8328645 | 0.58 |
ENSDART00000127494
|
nrg2b
|
neuregulin 2b |
| chr23_-_24582606 | 0.57 |
ENSDART00000129910
|
tmem240a
|
transmembrane protein 240a |
| chr18_+_45990394 | 0.57 |
ENSDART00000024068
|
mmp23bb
|
matrix metallopeptidase 23bb |
| chr6_+_6491013 | 0.57 |
ENSDART00000140827
|
bcl11ab
|
B cell CLL/lymphoma 11Ab |
| chr7_-_8490886 | 0.57 |
ENSDART00000159012
|
jac6
|
jacalin 6 |
| chr5_-_31712399 | 0.57 |
ENSDART00000141328
|
pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr23_+_34047413 | 0.56 |
ENSDART00000143933
ENSDART00000123925 ENSDART00000176139 |
chchd6a
|
coiled-coil-helix-coiled-coil-helix domain containing 6a |
| chr15_+_15779184 | 0.56 |
ENSDART00000156902
|
si:ch211-33e4.2
|
si:ch211-33e4.2 |
| chr21_-_25669820 | 0.56 |
ENSDART00000148236
|
tmem179b
|
transmembrane protein 179B |
| chr9_+_54686686 | 0.56 |
ENSDART00000066198
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr13_-_46991577 | 0.55 |
ENSDART00000114748
|
vip
|
vasoactive intestinal peptide |
| chr10_+_29199172 | 0.55 |
ENSDART00000148828
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr15_-_28247583 | 0.55 |
ENSDART00000112967
|
rilp
|
Rab interacting lysosomal protein |
| chr10_-_25860102 | 0.54 |
ENSDART00000080789
|
trpc4a
|
transient receptor potential cation channel, subfamily C, member 4a |
| chr17_-_24879003 | 0.54 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chr24_+_25259154 | 0.54 |
ENSDART00000171125
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr2_-_32501501 | 0.54 |
ENSDART00000181309
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr13_+_52061034 | 0.54 |
ENSDART00000170383
|
CABZ01089777.1
|
|
| chr16_-_32975951 | 0.54 |
ENSDART00000101969
ENSDART00000175149 |
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr5_+_72108241 | 0.53 |
ENSDART00000006606
|
fabp1a
|
fatty acid binding protein 1a, liver |
| chr5_+_32817688 | 0.53 |
ENSDART00000139472
|
crata
|
carnitine O-acetyltransferase a |
| chr5_-_30380593 | 0.52 |
ENSDART00000148039
|
snx19a
|
sorting nexin 19a |
| chr9_-_33749556 | 0.52 |
ENSDART00000149383
|
ndp
|
Norrie disease (pseudoglioma) |
| chr2_-_19128385 | 0.52 |
ENSDART00000169793
|
si:dkey-225f23.3
|
si:dkey-225f23.3 |
| chr3_-_21061931 | 0.51 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr2_-_32505091 | 0.51 |
ENSDART00000141884
ENSDART00000056639 |
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr19_-_27006764 | 0.51 |
ENSDART00000089540
|
sacm1la
|
SAC1 like phosphatidylinositide phosphatase a |
| chr16_+_26766423 | 0.50 |
ENSDART00000048036
|
gem
|
GTP binding protein overexpressed in skeletal muscle |
| chr17_+_36588281 | 0.50 |
ENSDART00000076115
|
htr1b
|
5-hydroxytryptamine (serotonin) receptor 1B |
| chr18_+_45796096 | 0.50 |
ENSDART00000087070
|
abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr5_-_41841892 | 0.49 |
ENSDART00000167089
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr7_-_61845282 | 0.49 |
ENSDART00000182586
|
HTRA3
|
HtrA serine peptidase 3 |
| chr19_+_33732188 | 0.49 |
ENSDART00000151192
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr2_+_39108339 | 0.49 |
ENSDART00000085675
|
clstn2
|
calsyntenin 2 |
| chr12_+_41991635 | 0.49 |
ENSDART00000186161
ENSDART00000192510 |
TCERG1L
|
transcription elongation regulator 1 like |
| chr7_-_20756013 | 0.48 |
ENSDART00000185259
|
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr12_+_25085751 | 0.47 |
ENSDART00000170466
|
gch2
|
GTP cyclohydrolase 2 |
| chr15_-_33933790 | 0.47 |
ENSDART00000165162
ENSDART00000182258 ENSDART00000183240 |
mag
|
myelin associated glycoprotein |
| chr3_-_30861177 | 0.47 |
ENSDART00000154811
|
shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr14_+_29581710 | 0.47 |
ENSDART00000188820
ENSDART00000193874 |
TENM2
|
si:dkey-34l15.2 |
| chr16_+_9713850 | 0.46 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr16_+_46453176 | 0.46 |
ENSDART00000132793
|
rpz3
|
rapunzel 3 |
| chr24_+_35947077 | 0.46 |
ENSDART00000173406
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr4_+_17336557 | 0.46 |
ENSDART00000111650
|
pmch
|
pro-melanin-concentrating hormone |
| chr14_+_25817628 | 0.46 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr16_-_29437373 | 0.45 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr25_+_10811551 | 0.45 |
ENSDART00000167730
|
anpepb
|
alanyl (membrane) aminopeptidase b |
| chr5_-_41841675 | 0.45 |
ENSDART00000141683
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr14_-_33936524 | 0.45 |
ENSDART00000112438
|
si:ch73-335m24.5
|
si:ch73-335m24.5 |
| chr16_+_24842662 | 0.45 |
ENSDART00000157333
|
si:dkey-79d12.6
|
si:dkey-79d12.6 |
| chr23_+_11669109 | 0.44 |
ENSDART00000091416
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr6_-_40899618 | 0.44 |
ENSDART00000153949
ENSDART00000021969 |
zgc:172271
|
zgc:172271 |
| chr22_-_18164671 | 0.44 |
ENSDART00000014057
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr23_-_40250886 | 0.44 |
ENSDART00000041912
|
tgm1l3
|
transglutaminase 1 like 3 |
| chr20_+_42668875 | 0.44 |
ENSDART00000048890
|
slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr1_-_201004 | 0.44 |
ENSDART00000188956
|
FO681286.1
|
|
| chr21_+_6290566 | 0.43 |
ENSDART00000161647
|
fnbp1b
|
formin binding protein 1b |
| chr23_+_16633951 | 0.43 |
ENSDART00000109537
ENSDART00000193323 |
snphb
|
syntaphilin b |
| chr25_-_1079417 | 0.43 |
ENSDART00000163134
|
FO907089.1
|
|
| chr23_+_44263986 | 0.43 |
ENSDART00000149194
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr20_+_2240278 | 0.43 |
ENSDART00000041250
|
tmem200a
|
transmembrane protein 200A |
| chr19_+_34230108 | 0.43 |
ENSDART00000141950
|
galnt12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 |
| chr8_-_44004135 | 0.43 |
ENSDART00000136269
|
rimbp2
|
RIMS binding protein 2 |
| chr22_+_30543437 | 0.43 |
ENSDART00000137983
|
si:dkey-103k4.1
|
si:dkey-103k4.1 |
| chr14_-_17068712 | 0.43 |
ENSDART00000170277
|
phox2bb
|
paired-like homeobox 2bb |
| chr23_+_11669337 | 0.43 |
ENSDART00000131355
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr22_-_282498 | 0.43 |
ENSDART00000182766
|
CABZ01079178.1
|
|
| chr7_-_47850702 | 0.43 |
ENSDART00000109511
|
si:ch211-186j3.6
|
si:ch211-186j3.6 |
| chr11_+_10541258 | 0.43 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr19_+_22216778 | 0.42 |
ENSDART00000052521
|
nfatc1
|
nuclear factor of activated T cells 1 |
| chr19_-_3488860 | 0.41 |
ENSDART00000172520
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr19_+_33732487 | 0.41 |
ENSDART00000010294
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr20_-_27390258 | 0.41 |
ENSDART00000010584
|
prph2l
|
peripherin 2, like |
| chr8_-_11146924 | 0.40 |
ENSDART00000126824
|
asb13b
|
ankyrin repeat and SOCS box containing 13b |
| chr19_+_12801940 | 0.40 |
ENSDART00000040073
|
mc5ra
|
melanocortin 5a receptor |
| chr12_+_17100021 | 0.40 |
ENSDART00000177923
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr20_-_25669813 | 0.40 |
ENSDART00000153118
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr20_+_27194833 | 0.40 |
ENSDART00000150072
|
si:dkey-85n7.8
|
si:dkey-85n7.8 |
| chr1_-_44413629 | 0.40 |
ENSDART00000192747
|
CT583626.1
|
|
| chr7_-_32833153 | 0.39 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr13_+_15816573 | 0.39 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of ikzf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.3 | 1.7 | GO:0021767 | mammillary body development(GO:0021767) social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.3 | 1.9 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.3 | 0.8 | GO:0042040 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.2 | 0.9 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 0.8 | GO:2000048 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 1.9 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.2 | 0.6 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.2 | 1.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 0.9 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.2 | 0.9 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 0.6 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.2 | 1.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.4 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 0.6 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.6 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 0.5 | GO:0070459 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.1 | 0.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 1.4 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.6 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 1.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.6 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.6 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.5 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.5 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.5 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 1.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.4 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 0.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 1.4 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 1.0 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.8 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 4.4 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.5 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.3 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 0.6 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.4 | GO:1901021 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.7 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.2 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.5 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.4 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.3 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0046222 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.0 | 0.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.7 | GO:0098868 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 | 0.2 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:1901004 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.0 | 0.2 | GO:0051883 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.4 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 1.1 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 1.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.3 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.4 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 1.0 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.4 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 1.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.7 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.3 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.9 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.5 | GO:0046849 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) bone remodeling(GO:0046849) |
| 0.0 | 1.1 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.7 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.7 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 1.4 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.7 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.4 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.7 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.0 | 0.4 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.0 | 1.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.3 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.0 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.1 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.1 | GO:0050938 | regulation of pigment cell differentiation(GO:0050932) regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.3 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.2 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.4 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 2.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.2 | GO:0044218 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.6 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.3 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 1.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 1.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 2.9 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.9 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 5.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.4 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.5 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.4 | 1.9 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.3 | 1.6 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.3 | 1.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 0.8 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.3 | 2.3 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 0.8 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 0.6 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.2 | 0.8 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 0.9 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 0.5 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 1.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.4 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.7 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.5 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 1.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.3 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.1 | 0.6 | GO:0015182 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.1 | 1.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.3 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.1 | 0.8 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.9 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.1 | 0.6 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.2 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.2 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.1 | 1.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.3 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.2 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 1.2 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 1.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.4 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.5 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.4 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.6 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.8 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.1 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.7 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 1.6 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 1.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0034979 | NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.8 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 1.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.8 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 0.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.6 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 1.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |