Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
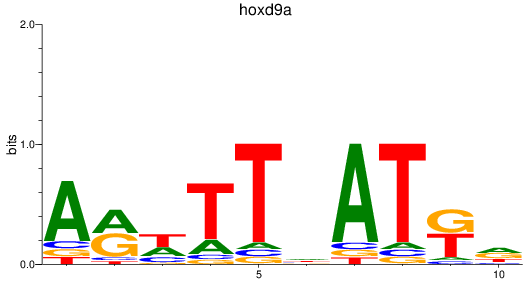
Results for hoxd9a
Z-value: 0.70
Transcription factors associated with hoxd9a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd9a
|
ENSDARG00000059274 | homeobox D9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd9a | dr11_v1_chr9_-_1965727_1965727 | -0.42 | 7.1e-02 | Click! |
Activity profile of hoxd9a motif
Sorted Z-values of hoxd9a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_21225750 | 2.11 |
ENSDART00000139213
|
zgc:153968
|
zgc:153968 |
| chr25_+_31227747 | 1.70 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr7_+_35068036 | 1.23 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr22_+_1049367 | 1.17 |
ENSDART00000065380
|
camk1ga
|
calcium/calmodulin-dependent protein kinase IGa |
| chr15_+_15390882 | 0.99 |
ENSDART00000062024
|
ca4b
|
carbonic anhydrase IV b |
| chr19_-_657439 | 0.88 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr20_-_33515890 | 0.88 |
ENSDART00000159987
|
si:dkey-65b13.13
|
si:dkey-65b13.13 |
| chr13_+_45709380 | 0.82 |
ENSDART00000192862
|
si:ch211-62a1.3
|
si:ch211-62a1.3 |
| chr20_-_52902693 | 0.73 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr3_+_1030479 | 0.72 |
ENSDART00000143449
|
zgc:172253
|
zgc:172253 |
| chr3_+_32142382 | 0.70 |
ENSDART00000133035
|
syt5a
|
synaptotagmin Va |
| chr13_-_37229245 | 0.70 |
ENSDART00000140923
|
si:dkeyp-77c8.5
|
si:dkeyp-77c8.5 |
| chr6_-_7776612 | 0.69 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr11_+_37201483 | 0.67 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr18_+_9615147 | 0.66 |
ENSDART00000160284
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr16_-_28658341 | 0.66 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr9_+_1139378 | 0.65 |
ENSDART00000170033
|
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr9_+_1138802 | 0.64 |
ENSDART00000191130
ENSDART00000187308 |
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr19_+_10339538 | 0.62 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr23_+_9035578 | 0.60 |
ENSDART00000140455
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr7_+_9189547 | 0.59 |
ENSDART00000169783
|
pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr23_+_37482727 | 0.59 |
ENSDART00000162737
|
agmat
|
agmatine ureohydrolase (agmatinase) |
| chr23_+_21544227 | 0.59 |
ENSDART00000140253
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr4_-_67969695 | 0.58 |
ENSDART00000190016
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr14_-_46617228 | 0.58 |
ENSDART00000161010
ENSDART00000171054 |
prom1a
|
prominin 1a |
| chr8_-_10048059 | 0.58 |
ENSDART00000138411
|
si:dkey-8e10.3
|
si:dkey-8e10.3 |
| chr2_+_45344070 | 0.57 |
ENSDART00000147245
|
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr10_-_2526526 | 0.57 |
ENSDART00000191726
ENSDART00000192767 |
CU856539.1
|
|
| chr12_+_1000323 | 0.54 |
ENSDART00000054363
|
si:ch1073-272o11.3
|
si:ch1073-272o11.3 |
| chr24_-_38574631 | 0.53 |
ENSDART00000154539
|
slc17a7b
|
solute carrier family 17 (vesicular glutamate transporter), member 7b |
| chr16_+_14707960 | 0.52 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr8_+_3820134 | 0.52 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr3_-_22214029 | 0.51 |
ENSDART00000154818
|
maptb
|
microtubule-associated protein tau b |
| chr10_+_23537576 | 0.51 |
ENSDART00000130515
|
CR847844.1
|
|
| chr6_+_40661703 | 0.51 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr22_+_37888249 | 0.51 |
ENSDART00000076082
|
fetub
|
fetuin B |
| chr19_+_4139065 | 0.51 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr21_+_21195487 | 0.51 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr19_-_10360188 | 0.48 |
ENSDART00000150933
|
si:ch211-232m10.6
|
si:ch211-232m10.6 |
| chr1_+_38858399 | 0.47 |
ENSDART00000165454
|
CU915762.1
|
|
| chr5_+_28830643 | 0.47 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr14_-_7141550 | 0.46 |
ENSDART00000172020
|
trpt1
|
tRNA phosphotransferase 1 |
| chr14_+_7892021 | 0.46 |
ENSDART00000161307
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr4_+_20954929 | 0.46 |
ENSDART00000143674
|
nav3
|
neuron navigator 3 |
| chr8_-_13184989 | 0.45 |
ENSDART00000135738
|
zgc:194990
|
zgc:194990 |
| chr25_-_30383617 | 0.44 |
ENSDART00000165342
|
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr18_-_8313686 | 0.44 |
ENSDART00000182187
|
mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr24_-_1985007 | 0.43 |
ENSDART00000189870
|
PARD3 (1 of many)
|
par-3 family cell polarity regulator |
| chr5_-_35201482 | 0.43 |
ENSDART00000166272
|
fcho2
|
FCH domain only 2 |
| chr19_+_1510971 | 0.43 |
ENSDART00000157721
|
SLC45A4 (1 of many)
|
solute carrier family 45 member 4 |
| chr5_-_41984298 | 0.43 |
ENSDART00000189535
|
ncor1
|
nuclear receptor corepressor 1 |
| chr23_-_14990865 | 0.42 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr3_+_60761811 | 0.41 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr22_-_10165446 | 0.41 |
ENSDART00000142012
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr23_+_41799748 | 0.41 |
ENSDART00000144257
|
pdyn
|
prodynorphin |
| chr20_+_18163821 | 0.41 |
ENSDART00000186507
|
aqp4
|
aquaporin 4 |
| chr9_-_14683574 | 0.40 |
ENSDART00000144022
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr13_+_7578111 | 0.40 |
ENSDART00000175431
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr5_-_24238733 | 0.39 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr21_-_20342096 | 0.39 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr15_-_3956906 | 0.39 |
ENSDART00000171202
|
clrn1
|
clarin 1 |
| chr13_+_6342410 | 0.39 |
ENSDART00000065228
|
csmd1a
|
CUB and Sushi multiple domains 1a |
| chr14_+_32430982 | 0.38 |
ENSDART00000017179
ENSDART00000123382 ENSDART00000075593 |
f9a
|
coagulation factor IXa |
| chr19_-_3167729 | 0.38 |
ENSDART00000110763
ENSDART00000145710 ENSDART00000074620 ENSDART00000105174 |
stm
|
starmaker |
| chr12_+_1286642 | 0.38 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr17_-_6730247 | 0.38 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr5_-_12701603 | 0.37 |
ENSDART00000091124
|
aifm3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr21_+_39963851 | 0.37 |
ENSDART00000144435
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr10_+_44903676 | 0.37 |
ENSDART00000158553
|
zgc:114173
|
zgc:114173 |
| chr3_-_46403778 | 0.37 |
ENSDART00000074422
|
cdip1
|
cell death-inducing p53 target 1 |
| chr24_-_35699595 | 0.37 |
ENSDART00000167990
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr25_+_14087045 | 0.36 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr2_-_4868002 | 0.36 |
ENSDART00000189835
|
tfr1a
|
transferrin receptor 1a |
| chr10_+_41765616 | 0.36 |
ENSDART00000170682
|
rnf34b
|
ring finger protein 34b |
| chr23_+_42819221 | 0.35 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr10_-_24371312 | 0.35 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr12_-_7807298 | 0.35 |
ENSDART00000191355
|
ank3b
|
ankyrin 3b |
| chr7_-_30087048 | 0.35 |
ENSDART00000112743
|
nmbb
|
neuromedin Bb |
| chr21_-_22115136 | 0.34 |
ENSDART00000134715
ENSDART00000089246 ENSDART00000139789 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr18_-_47662696 | 0.34 |
ENSDART00000184260
|
CABZ01073963.1
|
|
| chr2_-_39558643 | 0.34 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr18_+_9810642 | 0.33 |
ENSDART00000143965
|
CACNA2D1
|
si:dkey-266j7.2 |
| chr3_+_22442445 | 0.33 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr20_-_42100932 | 0.33 |
ENSDART00000191930
|
slc35f1
|
solute carrier family 35, member F1 |
| chr13_-_37620091 | 0.33 |
ENSDART00000135875
ENSDART00000193270 ENSDART00000018064 |
zgc:152791
|
zgc:152791 |
| chr3_-_38683327 | 0.32 |
ENSDART00000179781
|
mpp3a
|
membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) |
| chr18_+_2189211 | 0.32 |
ENSDART00000170827
|
ccpg1
|
cell cycle progression 1 |
| chr7_-_69636502 | 0.32 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr20_-_47188966 | 0.32 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr10_+_41765944 | 0.32 |
ENSDART00000171484
|
rnf34b
|
ring finger protein 34b |
| chr6_+_21536131 | 0.32 |
ENSDART00000113911
ENSDART00000188472 |
micall1a
|
MICAL-like 1a |
| chr17_+_30442518 | 0.32 |
ENSDART00000155264
|
lpin1
|
lipin 1 |
| chr5_-_12093618 | 0.31 |
ENSDART00000161542
|
lrrc74b
|
leucine rich repeat containing 74B |
| chr22_+_5752257 | 0.31 |
ENSDART00000143052
|
cox14
|
COX14 cytochrome c oxidase assembly factor |
| chr22_+_1028724 | 0.31 |
ENSDART00000149625
|
si:ch73-352p18.4
|
si:ch73-352p18.4 |
| chr14_+_39258569 | 0.30 |
ENSDART00000103298
|
diaph2
|
diaphanous-related formin 2 |
| chr19_+_7810028 | 0.30 |
ENSDART00000081592
ENSDART00000140719 |
aqp10b
|
aquaporin 10b |
| chr14_+_25464681 | 0.30 |
ENSDART00000067500
ENSDART00000187601 |
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr7_+_38510197 | 0.30 |
ENSDART00000173468
ENSDART00000100479 |
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr21_+_39941875 | 0.30 |
ENSDART00000190414
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr1_+_2321384 | 0.29 |
ENSDART00000157662
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr22_+_2409175 | 0.29 |
ENSDART00000141776
|
zgc:113220
|
zgc:113220 |
| chr23_-_1658980 | 0.29 |
ENSDART00000186227
|
CU693481.1
|
|
| chr21_-_20341836 | 0.28 |
ENSDART00000176689
|
rbp4l
|
retinol binding protein 4, like |
| chr11_-_4787083 | 0.28 |
ENSDART00000159425
|
FHIT
|
si:ch211-63i20.3 |
| chr22_+_2959879 | 0.28 |
ENSDART00000185354
ENSDART00000063545 |
impact
|
impact RWD domain protein |
| chr3_+_37083765 | 0.27 |
ENSDART00000125611
|
retreg3
|
reticulophagy regulator family member 3 |
| chr14_+_28492990 | 0.27 |
ENSDART00000160347
ENSDART00000187176 ENSDART00000088094 |
stag2b
|
stromal antigen 2b |
| chr10_+_16911951 | 0.27 |
ENSDART00000164933
|
UNC13B (1 of many)
|
unc-13 homolog B |
| chr14_+_45406299 | 0.27 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr9_+_52398531 | 0.27 |
ENSDART00000126215
|
dap1b
|
death associated protein 1b |
| chr2_+_33383227 | 0.27 |
ENSDART00000183068
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr3_+_35005062 | 0.27 |
ENSDART00000181163
|
prkcbb
|
protein kinase C, beta b |
| chr7_+_42935126 | 0.27 |
ENSDART00000157747
|
BX284696.1
|
|
| chr25_+_35019693 | 0.27 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr11_-_30611814 | 0.26 |
ENSDART00000089803
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr18_-_44908479 | 0.26 |
ENSDART00000169636
|
si:ch211-71n6.4
|
si:ch211-71n6.4 |
| chr11_-_26701611 | 0.26 |
ENSDART00000083010
|
acad9
|
acyl-CoA dehydrogenase family, member 9 |
| chr21_-_42831033 | 0.26 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr15_-_917274 | 0.26 |
ENSDART00000156624
|
si:dkey-77f5.15
|
si:dkey-77f5.15 |
| chr8_-_8446668 | 0.26 |
ENSDART00000132700
|
cdk16
|
cyclin-dependent kinase 16 |
| chr9_+_21993092 | 0.25 |
ENSDART00000059693
|
crygm7
|
crystallin, gamma M7 |
| chr5_-_23805387 | 0.25 |
ENSDART00000133805
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr9_+_33783061 | 0.25 |
ENSDART00000190193
|
BX572627.1
|
|
| chr14_-_45967981 | 0.25 |
ENSDART00000188062
|
macrod1
|
MACRO domain containing 1 |
| chr3_+_22327738 | 0.25 |
ENSDART00000055675
|
gh1
|
growth hormone 1 |
| chr19_+_47299212 | 0.24 |
ENSDART00000158262
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr23_-_26252103 | 0.24 |
ENSDART00000160873
|
magi3a
|
membrane associated guanylate kinase, WW and PDZ domain containing 3a |
| chr6_-_48473694 | 0.24 |
ENSDART00000154237
|
ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr18_-_46882056 | 0.24 |
ENSDART00000108574
|
gramd1bb
|
GRAM domain containing 1Bb |
| chr23_-_900795 | 0.23 |
ENSDART00000190517
ENSDART00000182849 ENSDART00000111456 ENSDART00000185430 |
rbm10
|
RNA binding motif protein 10 |
| chr19_-_31042570 | 0.23 |
ENSDART00000144337
ENSDART00000136213 ENSDART00000133101 ENSDART00000190949 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr3_+_13637383 | 0.23 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr19_-_40826024 | 0.23 |
ENSDART00000131471
|
calcr
|
calcitonin receptor |
| chr20_+_42978499 | 0.23 |
ENSDART00000138793
|
si:ch211-203k16.3
|
si:ch211-203k16.3 |
| chr23_-_41965557 | 0.23 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr7_-_26308098 | 0.23 |
ENSDART00000146440
ENSDART00000146935 ENSDART00000164627 |
zgc:77439
|
zgc:77439 |
| chr6_-_57539141 | 0.23 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr20_+_4793516 | 0.22 |
ENSDART00000053875
|
lgals8a
|
galectin 8a |
| chr3_+_22432069 | 0.22 |
ENSDART00000123054
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr18_-_36066087 | 0.22 |
ENSDART00000059352
ENSDART00000145177 |
exosc5
|
exosome component 5 |
| chr16_+_11660839 | 0.22 |
ENSDART00000193911
ENSDART00000143683 |
si:dkey-250k15.10
|
si:dkey-250k15.10 |
| chr4_+_71401321 | 0.22 |
ENSDART00000158076
|
si:ch211-76m11.7
|
si:ch211-76m11.7 |
| chr7_-_69983948 | 0.22 |
ENSDART00000185827
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr3_-_32472155 | 0.21 |
ENSDART00000156280
|
si:ch211-195b15.7
|
si:ch211-195b15.7 |
| chr13_+_25380432 | 0.21 |
ENSDART00000038524
|
gsto1
|
glutathione S-transferase omega 1 |
| chr19_+_2876106 | 0.21 |
ENSDART00000189309
|
hgh1
|
HGH1 homolog (S. cerevisiae) |
| chr7_-_24047316 | 0.21 |
ENSDART00000184799
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr18_+_44849809 | 0.21 |
ENSDART00000097328
|
arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr5_-_4239765 | 0.21 |
ENSDART00000180015
|
si:ch211-283g2.3
|
si:ch211-283g2.3 |
| chr12_-_4781801 | 0.21 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr9_+_23511913 | 0.20 |
ENSDART00000137759
|
cntnap5a
|
contactin associated protein-like 5a |
| chr20_+_41906960 | 0.20 |
ENSDART00000193460
|
cep85l
|
centrosomal protein 85, like |
| chr11_+_23760470 | 0.20 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr21_+_39941559 | 0.20 |
ENSDART00000189718
ENSDART00000160875 ENSDART00000135235 |
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr21_+_10018055 | 0.20 |
ENSDART00000163308
|
herc7
|
hect domain and RLD 7 |
| chr14_+_29975268 | 0.20 |
ENSDART00000172985
|
cyp4v7
|
cytochrome P450, family 4, subfamily V, polypeptide 7 |
| chr7_-_18730474 | 0.20 |
ENSDART00000170374
|
si:dkey-38l22.2
|
si:dkey-38l22.2 |
| chr17_+_20174812 | 0.20 |
ENSDART00000186628
|
si:ch211-248a14.8
|
si:ch211-248a14.8 |
| chr15_-_563877 | 0.19 |
ENSDART00000128032
|
cbln18
|
cerebellin 18 |
| chr8_-_52473054 | 0.19 |
ENSDART00000193658
|
phyhip
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr7_-_24046999 | 0.19 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr2_+_42072231 | 0.19 |
ENSDART00000084517
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr15_+_11825366 | 0.19 |
ENSDART00000190042
|
prkd2
|
protein kinase D2 |
| chr16_+_27383717 | 0.19 |
ENSDART00000132329
ENSDART00000136256 |
stx17
|
syntaxin 17 |
| chr6_-_53426773 | 0.19 |
ENSDART00000162791
|
mst1
|
macrophage stimulating 1 |
| chr3_-_49382896 | 0.18 |
ENSDART00000169115
|
si:ch73-167f10.1
|
si:ch73-167f10.1 |
| chr17_-_27419499 | 0.18 |
ENSDART00000186773
|
ythdf2
|
YTH N(6)-methyladenosine RNA binding protein 2 |
| chr4_+_14899720 | 0.18 |
ENSDART00000187456
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr21_+_21357309 | 0.18 |
ENSDART00000079634
ENSDART00000146102 |
kcnk12l
|
potassium channel, subfamily K, member 12 like |
| chr2_-_57344037 | 0.18 |
ENSDART00000148873
|
tcf3a
|
transcription factor 3a |
| chr7_-_26306546 | 0.18 |
ENSDART00000140817
|
zgc:77439
|
zgc:77439 |
| chr19_-_7272921 | 0.18 |
ENSDART00000102075
ENSDART00000132887 ENSDART00000130234 ENSDART00000193535 ENSDART00000136528 |
rxrba
|
retinoid x receptor, beta a |
| chr2_-_43191465 | 0.18 |
ENSDART00000025254
|
crema
|
cAMP responsive element modulator a |
| chr19_-_6083761 | 0.18 |
ENSDART00000151185
ENSDART00000143941 |
gsk3aa
|
glycogen synthase kinase 3 alpha a |
| chr6_-_58901552 | 0.17 |
ENSDART00000122003
|
mbd6
|
methyl-CpG binding domain protein 6 |
| chr4_+_77839900 | 0.17 |
ENSDART00000162243
|
si:dkey-207m2.4
|
si:dkey-207m2.4 |
| chr9_-_47472998 | 0.17 |
ENSDART00000134480
|
tns1b
|
tensin 1b |
| chr25_-_29087925 | 0.17 |
ENSDART00000171758
|
rpp25a
|
ribonuclease P and MRP subunit p25, a |
| chr2_+_11031360 | 0.17 |
ENSDART00000180020
ENSDART00000145093 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr12_+_24952902 | 0.17 |
ENSDART00000189086
ENSDART00000014868 |
calm3a
|
calmodulin 3a (phosphorylase kinase, delta) |
| chr4_+_14900042 | 0.17 |
ENSDART00000018261
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr21_+_39941184 | 0.17 |
ENSDART00000133604
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr6_-_31325400 | 0.17 |
ENSDART00000188869
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr13_-_30027730 | 0.16 |
ENSDART00000044009
|
scdb
|
stearoyl-CoA desaturase b |
| chr3_-_22213558 | 0.16 |
ENSDART00000178380
ENSDART00000187105 |
maptb
|
microtubule-associated protein tau b |
| chr2_+_10280645 | 0.16 |
ENSDART00000063996
|
gadd45aa
|
growth arrest and DNA-damage-inducible, alpha, a |
| chr8_-_22698651 | 0.16 |
ENSDART00000181411
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr4_+_75200467 | 0.16 |
ENSDART00000122593
|
CABZ01043955.1
|
|
| chr16_+_55168245 | 0.16 |
ENSDART00000171109
|
eomesb
|
eomesodermin homolog b |
| chr22_+_19538626 | 0.16 |
ENSDART00000189174
|
BX936298.2
|
|
| chr8_+_20950401 | 0.16 |
ENSDART00000136484
ENSDART00000131329 ENSDART00000144451 ENSDART00000144929 |
si:dkeyp-82a1.1
|
si:dkeyp-82a1.1 |
| chr14_-_2050057 | 0.16 |
ENSDART00000112875
|
PCDHB15
|
protocadherin beta 15 |
| chr6_+_32834007 | 0.15 |
ENSDART00000157353
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr25_+_8407892 | 0.15 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr13_-_31866276 | 0.15 |
ENSDART00000140164
|
syt14a
|
synaptotagmin XIVa |
| chr18_+_33580391 | 0.15 |
ENSDART00000189300
|
si:dkey-47k20.4
|
si:dkey-47k20.4 |
| chr10_+_40700311 | 0.15 |
ENSDART00000157650
ENSDART00000138342 |
taar19n
|
trace amine associated receptor 19n |
| chr8_-_13029297 | 0.15 |
ENSDART00000144305
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr2_-_11119303 | 0.15 |
ENSDART00000135450
ENSDART00000131836 |
cryz
|
crystallin, zeta (quinone reductase) |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd9a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.7 | GO:0072104 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 0.6 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.8 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.4 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 0.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.7 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.2 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.3 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 0.5 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.7 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.5 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.2 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 0.6 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.1 | 0.3 | GO:0010447 | response to acidic pH(GO:0010447) negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.4 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.1 | 0.2 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.2 | GO:0050996 | positive regulation of lipid catabolic process(GO:0050996) |
| 0.0 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.6 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.4 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.0 | 0.3 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.6 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.2 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.4 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0089700 | endocardial cushion formation(GO:0003272) protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.4 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.7 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.0 | 0.1 | GO:0034475 | tRNA catabolic process(GO:0016078) U1 snRNA 3'-end processing(GO:0034473) U4 snRNA 3'-end processing(GO:0034475) U5 snRNA 3'-end processing(GO:0034476) polyadenylation-dependent RNA catabolic process(GO:0043633) polyadenylation-dependent ncRNA catabolic process(GO:0043634) nuclear ncRNA surveillance(GO:0071029) nuclear polyadenylation-dependent rRNA catabolic process(GO:0071035) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) nuclear polyadenylation-dependent ncRNA catabolic process(GO:0071046) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.0 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.2 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.4 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.3 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.1 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.4 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.6 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 1.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.7 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 1.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.4 | GO:0032421 | stereocilium bundle(GO:0032421) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 1.3 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 0.7 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 0.4 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.2 | GO:0070186 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.1 | 0.2 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.2 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.2 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.1 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.0 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.5 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 0.3 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.7 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.6 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0015168 | water transmembrane transporter activity(GO:0005372) glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 1.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 4.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.0 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |