Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
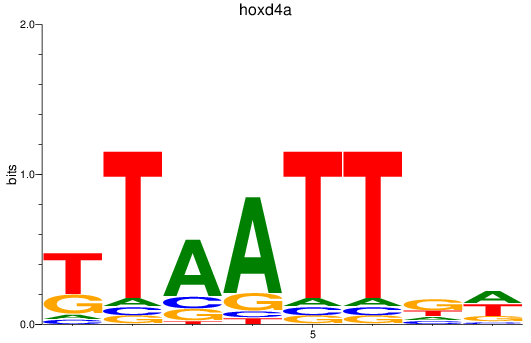
Results for hoxd4a
Z-value: 0.38
Transcription factors associated with hoxd4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd4a
|
ENSDARG00000059276 | homeobox D4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd4a | dr11_v1_chr9_-_1951144_1951144 | -0.31 | 1.9e-01 | Click! |
Activity profile of hoxd4a motif
Sorted Z-values of hoxd4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_26017360 | 0.92 |
ENSDART00000149466
|
prss59.2
|
protease, serine, 59, tandem duplicate 2 |
| chr3_+_21225750 | 0.64 |
ENSDART00000139213
|
zgc:153968
|
zgc:153968 |
| chr22_+_7439186 | 0.56 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr10_+_25726694 | 0.55 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr7_-_38698583 | 0.51 |
ENSDART00000173900
ENSDART00000126737 |
cd59
|
CD59 molecule (CD59 blood group) |
| chr10_+_17776981 | 0.40 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr15_+_45640906 | 0.37 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr4_+_7841627 | 0.36 |
ENSDART00000037997
|
ucmaa
|
upper zone of growth plate and cartilage matrix associated a |
| chr3_+_19460991 | 0.36 |
ENSDART00000169124
|
pde6ga
|
phosphodiesterase 6G, cGMP-specific, rod, gamma, paralog a |
| chr21_-_22689805 | 0.32 |
ENSDART00000157560
ENSDART00000110792 |
gig2e
|
grass carp reovirus (GCRV)-induced gene 2e |
| chr6_+_36381709 | 0.32 |
ENSDART00000004727
|
rhcgl1
|
Rh family, C glycoprotein, like 1 |
| chr15_-_34408777 | 0.32 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr7_-_20758825 | 0.31 |
ENSDART00000156717
ENSDART00000182629 ENSDART00000179801 |
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr8_+_3820134 | 0.31 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr6_-_46861676 | 0.31 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr16_-_13004166 | 0.30 |
ENSDART00000133735
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr3_+_24361096 | 0.30 |
ENSDART00000132387
|
pvalb6
|
parvalbumin 6 |
| chr3_-_4663602 | 0.29 |
ENSDART00000083532
|
slc25a38a
|
solute carrier family 25, member 38a |
| chr14_+_15620640 | 0.28 |
ENSDART00000188867
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr10_-_7756865 | 0.28 |
ENSDART00000114373
ENSDART00000125407 ENSDART00000016317 |
loxa
|
lysyl oxidase a |
| chr11_-_22303678 | 0.28 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr13_+_45709380 | 0.27 |
ENSDART00000192862
|
si:ch211-62a1.3
|
si:ch211-62a1.3 |
| chr1_+_10771457 | 0.26 |
ENSDART00000027050
ENSDART00000137185 |
cnga3b
|
cyclic nucleotide gated channel alpha 3b |
| chr8_+_52284075 | 0.26 |
ENSDART00000098439
|
ube2d4
|
ubiquitin-conjugating enzyme E2D 4 (putative) |
| chr2_-_39558643 | 0.26 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr20_+_106481 | 0.26 |
ENSDART00000148834
|
si:ch1073-155h21.2
|
si:ch1073-155h21.2 |
| chr25_-_10503043 | 0.25 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr11_+_44804685 | 0.25 |
ENSDART00000163660
|
strn
|
striatin, calmodulin binding protein |
| chr4_+_15944245 | 0.24 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr8_+_26230060 | 0.23 |
ENSDART00000155529
ENSDART00000140798 |
wdr6
|
WD repeat domain 6 |
| chr20_+_18551657 | 0.23 |
ENSDART00000147001
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr3_-_49382896 | 0.23 |
ENSDART00000169115
|
si:ch73-167f10.1
|
si:ch73-167f10.1 |
| chr22_-_10470663 | 0.23 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr20_+_23173710 | 0.23 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr2_+_3201345 | 0.22 |
ENSDART00000130349
|
wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr15_-_21877726 | 0.22 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr12_+_20352400 | 0.22 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr13_+_33368140 | 0.21 |
ENSDART00000033848
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr7_+_33145529 | 0.21 |
ENSDART00000052387
|
itln2
|
intelectin 2 |
| chr19_+_43297546 | 0.20 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr4_+_20536120 | 0.20 |
ENSDART00000125685
|
zmp:0000000650
|
zmp:0000000650 |
| chr16_+_14707960 | 0.19 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr20_-_14206698 | 0.19 |
ENSDART00000082441
|
dlgap2b
|
discs, large (Drosophila) homolog-associated protein 2b |
| chr23_-_1017605 | 0.19 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr17_-_4252221 | 0.19 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr20_+_10544100 | 0.19 |
ENSDART00000113927
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr7_+_69905307 | 0.19 |
ENSDART00000035907
|
sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr15_-_30816370 | 0.18 |
ENSDART00000142982
ENSDART00000050649 ENSDART00000136901 ENSDART00000100194 |
msi2b
|
musashi RNA-binding protein 2b |
| chr5_-_68795063 | 0.18 |
ENSDART00000016307
|
her1
|
hairy-related 1 |
| chr6_+_39098397 | 0.18 |
ENSDART00000003716
ENSDART00000188655 |
prss60.2
|
protease, serine, 60.2 |
| chr7_-_73851280 | 0.18 |
ENSDART00000190053
|
FP236812.3
|
|
| chr7_-_20582842 | 0.18 |
ENSDART00000169750
ENSDART00000111719 |
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr5_-_1869982 | 0.17 |
ENSDART00000055878
|
rcl1
|
RNA terminal phosphate cyclase-like 1 |
| chr13_-_9318891 | 0.17 |
ENSDART00000137364
|
si:dkey-33c12.3
|
si:dkey-33c12.3 |
| chr23_-_14990865 | 0.17 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr13_-_37631092 | 0.17 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr22_+_18166660 | 0.16 |
ENSDART00000105432
|
borcs8
|
BLOC-1 related complex subunit 8 |
| chr7_+_65309738 | 0.16 |
ENSDART00000169228
|
vat1l
|
vesicle amine transport 1-like |
| chr18_+_13077800 | 0.16 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr13_-_856525 | 0.16 |
ENSDART00000143356
|
TMEM14A
|
zgc:163080 |
| chr22_+_8753365 | 0.16 |
ENSDART00000106086
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr20_+_36628059 | 0.16 |
ENSDART00000062898
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr10_+_21522545 | 0.16 |
ENSDART00000131097
|
BX957322.1
|
|
| chr7_+_12927944 | 0.15 |
ENSDART00000027329
|
fth1a
|
ferritin, heavy polypeptide 1a |
| chr19_-_47527093 | 0.15 |
ENSDART00000171379
ENSDART00000171140 ENSDART00000114886 |
scg5
|
secretogranin V |
| chr18_+_7504691 | 0.15 |
ENSDART00000169075
|
neto2b
|
neuropilin (NRP) and tolloid (TLL)-like 2b |
| chr9_-_19762801 | 0.15 |
ENSDART00000146841
|
si:ch211-141e20.2
|
si:ch211-141e20.2 |
| chr12_+_47698356 | 0.15 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr13_+_2861265 | 0.15 |
ENSDART00000170602
ENSDART00000171687 |
XPO5
|
si:ch211-233m11.2 |
| chr9_-_464737 | 0.14 |
ENSDART00000170472
|
si:dkey-11f4.16
|
si:dkey-11f4.16 |
| chr7_+_22657566 | 0.14 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr7_+_5910467 | 0.14 |
ENSDART00000173232
|
si:ch211-113a14.22
|
si:ch211-113a14.22 |
| chr19_+_22074468 | 0.14 |
ENSDART00000136294
ENSDART00000090476 |
atp9b
|
ATPase phospholipid transporting 9B |
| chr15_-_20400423 | 0.14 |
ENSDART00000081290
ENSDART00000156813 ENSDART00000192427 |
rnaset2l
|
ribonuclease T2, like |
| chr24_-_35707552 | 0.14 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr21_-_13123176 | 0.14 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr7_-_6446172 | 0.14 |
ENSDART00000173212
|
hist1h2a11
|
histone cluster 1 H2A family member 11 |
| chr16_-_17541890 | 0.14 |
ENSDART00000131328
|
clcn1b
|
chloride channel, voltage-sensitive 1b |
| chr11_-_30611814 | 0.13 |
ENSDART00000089803
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr2_+_2181709 | 0.13 |
ENSDART00000092763
ENSDART00000148553 |
ccdc13
|
coiled-coil domain containing 13 |
| chr15_+_42397125 | 0.13 |
ENSDART00000169751
|
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr6_+_39279831 | 0.13 |
ENSDART00000155088
|
ankrd33ab
|
ankyrin repeat domain 33Ab |
| chr2_-_23677422 | 0.13 |
ENSDART00000079131
|
cdyl
|
chromodomain protein, Y-like |
| chr11_+_15878343 | 0.13 |
ENSDART00000167191
ENSDART00000171862 ENSDART00000163992 ENSDART00000170065 |
pank4
|
pantothenate kinase 4 |
| chr2_-_24398324 | 0.13 |
ENSDART00000165226
|
zgc:154006
|
zgc:154006 |
| chr16_+_29509133 | 0.13 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr11_-_28911172 | 0.12 |
ENSDART00000168493
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr23_+_2760573 | 0.12 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr4_+_14900042 | 0.12 |
ENSDART00000018261
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr3_-_31258459 | 0.12 |
ENSDART00000177928
|
LO017965.1
|
|
| chr4_-_19921410 | 0.12 |
ENSDART00000184359
ENSDART00000179965 |
BX323820.1
|
|
| chr8_+_28467893 | 0.12 |
ENSDART00000189724
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr20_-_48604621 | 0.12 |
ENSDART00000161769
ENSDART00000157871 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr8_-_44004135 | 0.12 |
ENSDART00000136269
|
rimbp2
|
RIMS binding protein 2 |
| chr8_+_28065803 | 0.12 |
ENSDART00000178481
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr21_+_21195487 | 0.12 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr18_+_7073130 | 0.12 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr4_+_16787488 | 0.12 |
ENSDART00000143006
|
golt1ba
|
golgi transport 1Ba |
| chr6_+_21395051 | 0.12 |
ENSDART00000017774
|
cacng5a
|
calcium channel, voltage-dependent, gamma subunit 5a |
| chr2_+_37815687 | 0.11 |
ENSDART00000166352
|
sdr39u1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr6_+_21992820 | 0.11 |
ENSDART00000147507
|
thumpd3
|
THUMP domain containing 3 |
| chr24_-_13364311 | 0.11 |
ENSDART00000183808
|
KCNB2
|
si:dkey-192j17.1 |
| chr18_-_16392229 | 0.11 |
ENSDART00000189031
|
mgat4c
|
mgat4 family, member C |
| chr20_-_16171297 | 0.11 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr25_-_24248000 | 0.10 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr19_+_4139065 | 0.10 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr10_-_41980797 | 0.10 |
ENSDART00000076575
|
rhof
|
ras homolog family member F |
| chr1_-_18811517 | 0.10 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr9_+_29616854 | 0.10 |
ENSDART00000033902
ENSDART00000143493 |
phf11
|
PHD finger protein 11 |
| chr7_-_27038488 | 0.10 |
ENSDART00000052731
ENSDART00000191382 |
nucb2a
|
nucleobindin 2a |
| chr18_-_5392667 | 0.10 |
ENSDART00000169896
|
CABZ01084603.1
|
|
| chr25_-_13839743 | 0.10 |
ENSDART00000158780
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr11_+_1608348 | 0.10 |
ENSDART00000162438
|
si:dkey-40c23.3
|
si:dkey-40c23.3 |
| chr9_-_36924388 | 0.10 |
ENSDART00000059756
|
ralba
|
v-ral simian leukemia viral oncogene homolog Ba (ras related) |
| chr24_-_35269991 | 0.10 |
ENSDART00000185424
|
sntg1
|
syntrophin, gamma 1 |
| chr19_-_1855368 | 0.09 |
ENSDART00000029646
|
rplp1
|
ribosomal protein, large, P1 |
| chr25_+_34888886 | 0.09 |
ENSDART00000035245
|
spire2
|
spire-type actin nucleation factor 2 |
| chr17_+_44697604 | 0.09 |
ENSDART00000156625
|
pgfb
|
placental growth factor b |
| chr10_+_39893439 | 0.09 |
ENSDART00000003435
|
smfn
|
small fragment nuclease |
| chr23_+_19590006 | 0.09 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr19_-_3167729 | 0.09 |
ENSDART00000110763
ENSDART00000145710 ENSDART00000074620 ENSDART00000105174 |
stm
|
starmaker |
| chr5_+_26847190 | 0.09 |
ENSDART00000076742
|
imp4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr24_-_37640705 | 0.09 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr18_+_3572314 | 0.09 |
ENSDART00000169814
ENSDART00000157819 |
serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr13_-_49585385 | 0.09 |
ENSDART00000165868
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr14_-_3070613 | 0.09 |
ENSDART00000193729
ENSDART00000090196 |
slc35a4
|
solute carrier family 35, member A4 |
| chr20_-_54924593 | 0.08 |
ENSDART00000151522
|
si:dkey-15f23.1
|
si:dkey-15f23.1 |
| chr18_-_33979693 | 0.08 |
ENSDART00000021215
|
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr7_-_30280934 | 0.08 |
ENSDART00000126741
|
serf2
|
small EDRK-rich factor 2 |
| chr7_-_2129076 | 0.08 |
ENSDART00000182385
|
si:cabz01007807.1
|
si:cabz01007807.1 |
| chr1_-_46875493 | 0.08 |
ENSDART00000115081
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr7_+_48761875 | 0.08 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr7_-_24699985 | 0.08 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr13_+_2625150 | 0.08 |
ENSDART00000164177
|
plpp4
|
phospholipid phosphatase 4 |
| chr4_+_20576857 | 0.08 |
ENSDART00000125340
|
BX248410.2
|
|
| chr5_+_24882633 | 0.08 |
ENSDART00000111302
|
rhbdd3
|
rhomboid domain containing 3 |
| chr12_+_18681477 | 0.07 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr21_-_42831033 | 0.07 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr14_-_31489410 | 0.07 |
ENSDART00000018347
|
cab39l1
|
calcium binding protein 39, like 1 |
| chr25_+_14165447 | 0.07 |
ENSDART00000145387
|
shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr24_+_21621654 | 0.07 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr5_-_10239079 | 0.07 |
ENSDART00000132739
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr23_-_16485190 | 0.07 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr9_+_30720048 | 0.07 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr4_-_14915268 | 0.07 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr18_+_30508729 | 0.07 |
ENSDART00000185140
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr2_+_9755422 | 0.07 |
ENSDART00000018524
|
pcyt1aa
|
phosphate cytidylyltransferase 1, choline, alpha a |
| chr22_+_16555939 | 0.07 |
ENSDART00000012604
|
pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr21_+_4256291 | 0.07 |
ENSDART00000148138
|
lrrc8aa
|
leucine rich repeat containing 8 VRAC subunit Aa |
| chr7_-_17570923 | 0.06 |
ENSDART00000188476
ENSDART00000080624 |
nitr5
|
novel immune-type receptor 5 |
| chr13_+_30692669 | 0.06 |
ENSDART00000187818
|
CR762483.1
|
|
| chr24_+_15020402 | 0.06 |
ENSDART00000148102
|
dok6
|
docking protein 6 |
| chr21_-_19314618 | 0.06 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr2_+_54389750 | 0.06 |
ENSDART00000189236
|
rab12
|
RAB12, member RAS oncogene family |
| chr11_+_1602916 | 0.06 |
ENSDART00000184434
ENSDART00000112597 ENSDART00000192165 |
si:dkey-40c23.2
si:dkey-40c23.3
|
si:dkey-40c23.2 si:dkey-40c23.3 |
| chr9_+_31534601 | 0.06 |
ENSDART00000133094
|
si:ch211-168k14.2
|
si:ch211-168k14.2 |
| chr6_-_39270851 | 0.06 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr18_+_16192083 | 0.06 |
ENSDART00000133042
|
lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
| chr12_+_28799988 | 0.06 |
ENSDART00000022724
|
pnpo
|
pyridoxamine 5'-phosphate oxidase |
| chr14_+_22022441 | 0.06 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr3_-_39259919 | 0.06 |
ENSDART00000124845
|
nmt1a
|
N-myristoyltransferase 1a |
| chr7_+_19495379 | 0.06 |
ENSDART00000180514
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr25_+_8407892 | 0.05 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr14_-_7137808 | 0.05 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr4_-_1908179 | 0.05 |
ENSDART00000139586
|
ano6
|
anoctamin 6 |
| chr13_+_40034176 | 0.05 |
ENSDART00000189797
|
golga7bb
|
golgin A7 family, member Bb |
| chr23_+_40460333 | 0.05 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr2_+_38370535 | 0.05 |
ENSDART00000179614
|
psmb11a
|
proteasome subunit beta 11a |
| chr18_+_44849809 | 0.05 |
ENSDART00000097328
|
arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr9_+_48219111 | 0.05 |
ENSDART00000111225
ENSDART00000145972 |
ccdc173
|
coiled-coil domain containing 173 |
| chr19_+_24872159 | 0.05 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr2_+_36121373 | 0.05 |
ENSDART00000187002
|
CT867973.2
|
|
| chr11_-_41220794 | 0.05 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr12_+_10062953 | 0.05 |
ENSDART00000148689
|
slc4a1b
|
solute carrier family 4 (anion exchanger), member 1b (Diego blood group) |
| chr4_-_49582108 | 0.05 |
ENSDART00000154999
|
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr24_+_26134209 | 0.04 |
ENSDART00000038824
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr2_+_16160906 | 0.04 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr2_+_45362016 | 0.04 |
ENSDART00000139154
|
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr14_+_25986895 | 0.04 |
ENSDART00000149087
|
slc36a1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr2_+_23677179 | 0.04 |
ENSDART00000153918
|
oxsr1a
|
oxidative stress responsive 1a |
| chr25_+_18563476 | 0.04 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr2_+_20605186 | 0.04 |
ENSDART00000128505
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr17_+_21477892 | 0.04 |
ENSDART00000155309
|
pla2g4f.2
|
phospholipase A2, group IVF, tandem duplicate 2 |
| chr14_-_35462148 | 0.04 |
ENSDART00000045809
|
srpx2
|
sushi-repeat containing protein, X-linked 2 |
| chr17_-_13070602 | 0.04 |
ENSDART00000188311
ENSDART00000193428 |
CU469462.1
|
|
| chr18_-_30020879 | 0.04 |
ENSDART00000162086
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr19_+_45962016 | 0.04 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr9_+_34380299 | 0.04 |
ENSDART00000131705
|
lamp1
|
lysosomal-associated membrane protein 1 |
| chr17_+_25481466 | 0.03 |
ENSDART00000139451
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr11_-_40101246 | 0.03 |
ENSDART00000161083
|
tnfrsf9b
|
tumor necrosis factor receptor superfamily, member 9b |
| chr7_-_3429874 | 0.03 |
ENSDART00000132330
|
si:ch211-285c6.1
|
si:ch211-285c6.1 |
| chr15_+_5339056 | 0.03 |
ENSDART00000174319
|
CABZ01018874.2
|
|
| chr14_+_4807207 | 0.03 |
ENSDART00000167145
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr5_-_66768121 | 0.03 |
ENSDART00000141095
|
FERMT3 (1 of many)
|
im:7154036 |
| chr2_+_16696052 | 0.03 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr4_+_17671164 | 0.03 |
ENSDART00000040445
|
gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr14_-_33481428 | 0.02 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr14_-_899170 | 0.02 |
ENSDART00000165211
ENSDART00000031992 |
rgs14a
|
regulator of G protein signaling 14a |
| chr16_-_15988320 | 0.02 |
ENSDART00000160883
|
CABZ01060453.1
|
|
| chr15_-_28082310 | 0.02 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.1 | 0.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.4 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 | 0.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.1 | GO:0097623 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.2 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.4 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.1 | GO:0045778 | positive regulation of bone mineralization(GO:0030501) positive regulation of ossification(GO:0045778) positive regulation of biomineral tissue development(GO:0070169) |
| 0.0 | 0.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.4 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.4 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.5 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 0.1 | GO:0042823 | water-soluble vitamin biosynthetic process(GO:0042364) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.3 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.0 | GO:0015808 | L-alanine transport(GO:0015808) proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.1 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 0.1 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.0 | 0.2 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.2 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.2 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.2 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.2 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.3 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |